- Search by keyword
- Search by citation
Page 1 of 56

The roles of cell wall polysaccharides in response to waterlogging stress in Brassica napus L. root
Brassica napus L. ( B. napus ) is susceptible to waterlogging stress during different cultivation periods. Therefore, it is crucial to enhance the resistance to waterlogging stress to achieve a high and stable yiel...
- View Full Text
Phylogeny and evolution of hemipteran insects based on expanded genomic and transcriptomic data
Hemiptera is the fifth species-rich order of insects and the most species-rich order of hemimetabolous insects, including numerous insect species that are of agricultural or medical significance. Despite much ...
Genome binding properties of Zic transcription factors underlie their changing functions during neuronal maturation
The Zic family of transcription factors (TFs) promote both proliferation and maturation of cerebellar granule neurons (CGNs), raising the question of how a single, constitutively expressed TF family can suppor...
Identification of a novel DNA oxidative damage repair pathway, requiring the ubiquitination of the histone variant macroH2A1.1
The histone variant macroH2A (mH2A), the most deviant variant, is about threefold larger than the conventional histone H2A and consists of a histone H2A-like domain fused to a large Non-Histone Region responsi...
Introgression drives adaptation to the plateau environment in a subterranean rodent
Introgression has repeatedly been shown to play an important role in the adaptation of species to extreme environments, yet how introgression enables rodents with specialized subterranean lifestyle to acclimat...
Alterations of pleiotropic neuropeptide-receptor gene couples in Cetacea
Habitat transitions have considerable consequences in organism homeostasis, as they require the adjustment of several concurrent physiological compartments to maintain stability and adapt to a changing environ...
Phylogenomics analysis of Scutellaria (Lamiaceae) of the world
Scutellaria , a sub-cosmopolitan genus, stands as one of the Lamiaceae family’s largest genera, encompassing approximately 500 species found in both temperate and tropical montane regions. Recognized for its signi...
PmLBD3 links auxin and brassinosteroid signalling pathways on dwarfism in Prunus mume
Grafting with dwarf rootstock is an efficient method to control plant height in fruit production. However, the molecular mechanism remains unclear. Our previous study showed that plants with Prunus mume (mume) ro...
You talkin’ to me? Functional breed selection may have fundamentally influenced dogs’ sensitivity to human verbal communicative cues
The ability to learn from humans via observation was considered to be equally present across properly socialized dogs. We showed recently that cooperative working breeds learned from a human demonstrator more ...

MvGraphDTA: multi-view-based graph deep model for drug-target affinity prediction by introducing the graphs and line graphs
Accurately identifying drug-target affinity (DTA) plays a pivotal role in drug screening, design, and repurposing in pharmaceutical industry. It not only reduces the time, labor, and economic costs associated ...
Flow cytometric analysis for Ki67 assessment in formalin-fixed paraffin-embedded breast cancer tissue
Pathologists commonly employ the Ki67 immunohistochemistry labelling index (LI) when deciding appropriate therapeutic strategies for patients with breast cancer. However, despite several attempts at standardiz...
Foxg1 regulates translation of neocortical neuronal genes, including the main NMDA receptor subunit gene, Grin1
Mainly known as a transcription factor patterning the rostral brain and governing its histogenesis, FOXG1 has been also detected outside the nucleus; however, biological meaning of that has been only partially...
Phospho-code of a conserved transcriptional factor underpins fungal virulence
Cell wall integrity (CWI) is crucial for fungal growth, pathogenesis, and adaptation to extracellular environments. Calcofluor white (CFW) is a cell wall perturbant that inhibits fungal growth, yet little is k...
Non-human peptides revealed in blood reflect the composition of intestinal microbiota
The previously underestimated effects of commensal gut microbiota on the human body are increasingly being investigated using omics. The discovery of active molecules of interaction between the microbiota and ...
G-quadruplexes as pivotal components of cis -regulatory elements in the human genome
Cis -regulatory elements (CREs) are crucial for regulating gene expression, and G-quadruplexes (G4s), as prototypal non-canonical DNA structures, may play a role in this regulation. However, the relationship betwe...
Casein kinase 1α mediates estradiol secretion via CYP19A1 expression in mouse ovarian granulosa cells
Casein kinase 1α (CK1α), expressed in both ovarian germ and somatic cells, is involved in the initial meiosis and primordial follicle formation of mouse oocytes. Using in vitro and in vivo experiments in this ...
The impact of filamentous plant pathogens on the host microbiota
When a pathogen invades a plant, it encounters a diverse microbiota with some members contributing to the health and growth of the plant host. So far, the relevance of interactions between pathogens and the pl...
Host and venom evolution in parasitoid wasps: does independently adapting to the same host shape the evolution of the venom gland transcriptome?
Venoms have repeatedly evolved over 100 occasions throughout the animal tree of life, making them excellent systems for exploring convergent evolutionary novelty. Growing evidence supports that venom evolution...
Intricate response dynamics enhances stimulus discrimination in the resource-limited C. elegans chemosensory system
Sensory systems evolved intricate designs to accurately encode perplexing environments. However, this encoding task may become particularly challenging for animals harboring a small number of sensory neurons. ...
Identification of microbe–disease signed associations via multi-scale variational graph autoencoder based on signed message propagation
Plenty of clinical and biomedical research has unequivocally highlighted the tremendous significance of the human microbiome in relation to human health. Identifying microbes associated with diseases is crucia...
Lipases are differentially regulated by hormones to maintain free fatty acid homeostasis for insect brain development
Free fatty acids (FFAs) play vital roles as energy sources and substrates in organisms; however, the molecular mechanism regulating the homeostasis of FFA levels in various circumstances, such as feeding and n...
Complex evolutionary patterns within the tubulin gene family of ciliates, unicellular eukaryotes with diverse microtubular structures
Tubulins are major components of the eukaryotic cytoskeletons that are crucial in many cellular processes. Ciliated protists comprise one of the oldest eukaryotic lineages possessing cilia over their cell surf...
CRY1 is involved in the take-off behaviour of migratory Cnaphalocrocis medinalis individuals
Numerous insect species undertake long-distance migrations on an enormous scale, with great implications for ecosystems. Given that take-off is the point where it all starts, whether and how the external light...
Life on a leaf: the epiphyte to pathogen continuum and interplay in the phyllosphere
Epiphytic microbes are those that live for some or all of their life cycle on the surface of plant leaves. Leaf surfaces are a topologically complex, physicochemically heterogeneous habitat that is home to ext...
COMSE: analysis of single-cell RNA-seq data using community detection-based feature selection
Single-cell RNA sequencing enables studying cells individually, yet high gene dimensions and low cell numbers challenge analysis. And only a subset of the genes detected are involved in the biological processe...
Monoamine oxidases activity maintains endometrial monoamine homeostasis and participates in embryo implantation and development
Monoamine oxidases (MAOs) is an enzyme that catalyzes the deamination of monoamines. The current research on this enzyme is focused on its role in neuropsychiatric, neurodevelopmental, and neurodegenerative di...
De novo genome assembly of white clover ( Trifolium repens L.) reveals the role of copy number variation in rapid environmental adaptation
White clover ( Trifolium repens ) is a globally important perennial forage legume. This species also serves as an eco-evolutionary model system for studying within-species chemical defense variation; it features a ...
Highly regenerative species-specific genes improve age-associated features in the adult Drosophila midgut
The remarkable regenerative abilities observed in planarians and cnidarians are closely linked to the active proliferation of adult stem cells and the precise differentiation of their progeny, both of which ty...
Diversely evolved xibalbin variants from remipede venom inhibit potassium channels and activate PKA-II and Erk1/2 signaling
The identification of novel toxins from overlooked and taxonomically exceptional species bears potential for various pharmacological applications. The remipede Xibalbanus tulumensis , an underwater cave-dwelling c...
Somatostatin signalling coordinates energy metabolism allocation to reproduction in zebrafish
Energy allocation between growth and reproduction determines puberty onset and fertility. In mammals, peripheral hormones such as leptin, insulin and ghrelin signal metabolic information to the higher centres ...
Single-mitochondrion sequencing uncovers distinct mutational patterns and heteroplasmy landscape in mouse astrocytes and neurons
Mitochondrial (mt) heteroplasmy can cause adverse biological consequences when deleterious mtDNA mutations accumulate disrupting “normal” mt-driven processes and cellular functions. To investigate the heteropl...
From birth to bite: the evolutionary ecology of India's medically most important snake venoms
Snake venoms can exhibit remarkable inter- and intraspecific variation. While diverse ecological and environmental factors are theorised to explain this variation, only a handful of studies have attempted to u...
Keratinocytes drive the epithelial hyperplasia key to sea lice resistance in coho salmon
Salmonid species have followed markedly divergent evolutionary trajectories in their interactions with sea lice. While sea lice parasitism poses significant economic, environmental, and animal welfare challeng...
Hierarchical lncRNA regulatory network in early-onset severe preeclampsia
Recent studies have shown that several long non-coding RNAs (lncRNAs) in the placenta are associated with preeclampsia (PE). However, the extent to which lncRNAs may contribute to the pathological progression ...
Plancitoxin-1 mediates extracellular trap evasion by the parasitic helminth Trichinella spiralis
Trichinella spiralis ( T. spiralis ) is a parasitic helminth that causes a globally prevalent neglected zoonotic disease, and worms at different developmental stages (muscle larvae, adult worms, newborn larvae) ind...
GSRF-DTI: a framework for drug-target interaction prediction based on a drug-target pair network and representation learning on a large graph
Identification of potential drug-target interactions (DTIs) with high accuracy is a key step in drug discovery and repositioning, especially concerning specific drug targets. Traditional experimental methods f...
Author Correction: PCYT1A deficiency disturbs fatty acid metabolism and induces ferroptosis in the mouse retina
The original article was published in BMC Biology 2024 22 :134
Endosomal protein DENND10/FAM45A integrates extracellular vesicle release with cancer cell migration
Mounting evidence shows that tumor-derived extracellular vesicles (EVs) are critical constituents in the tumor microenvironment. The composition and function of EVs often change during cancer progression. Howe...
AtSNU13 modulates pre-mRNA splicing of RBOHD and ALD1 to regulate plant immunity
Pre-mRNA splicing is a significant step for post-transcriptional modifications and functions in a wide range of physiological processes in plants. Human NHP2L binds to U4 snRNA during spliceosome assembly; it ...
Predicting intercellular communication based on metabolite-related ligand-receptor interactions with MRCLinkdb
Metabolite-associated cell communications play critical roles in maintaining human biological function. However, most existing tools and resources focus only on ligand-receptor interaction pairs where both par...
RNA 2'-O-methylation promotes persistent R-loop formation and AID-mediated IgH class switch recombination
RNA–DNA hybrids or R-loops are associated with deleterious genomic instability and protective immunoglobulin class switch recombination (CSR). However, the underlying phenomenon regulating the two contrasting ...
SNMP1 is critical for sensitive detection of the desert locust aromatic courtship inhibition pheromone phenylacetonitrile
Accurate detection of pheromones is crucial for chemical communication and reproduction in insects. In holometabolous flies and moths, the sensory neuron membrane protein 1 (SNMP1) is essential for detecting l...
Natural variation in yeast reveals multiple paths for acquiring higher stress resistance
Organisms frequently experience environmental stresses that occur in predictable patterns and combinations. For wild Saccharomyces cerevisiae yeast growing in natural environments, cells may experience high osmot...
Microbial diversity and ecological complexity emerging from environmental variation and horizontal gene transfer in a simple mathematical model
Microbiomes are generally characterized by high diversity of coexisting microbial species and strains, and microbiome composition typically remains stable across a broad range of conditions. However, under fix...
DNA methylation of exercise-responsive genes differs between trained and untrained men
Physical activity is well known for its multiple health benefits and although the knowledge of the underlying molecular mechanisms is increasing, our understanding of the role of epigenetics in long-term train...
Disturbed intracellular folate homeostasis impairs autophagic flux and increases hepatocytic lipid accumulation
Metabolic associated fatty liver disease (MAFLD), a prevalent liver disorder affecting one-third of the global population, encompasses a spectrum ranging from fatty liver to severe hepatic steatosis. Both gene...
Genomics-based identification of a cold adapted clade in Deinococcus
Microbes in the cold polar and alpine environments play a critical role in feedbacks that amplify the effects of climate change. Defining the cold adapted ecotype is one of the prerequisites for understanding ...
CRISPR-based genome editing of a diurnal rodent, Nile grass rat ( Arvicanthis niloticus)
Diurnal and nocturnal mammals have evolved distinct pathways to optimize survival for their chronotype-specific lifestyles. Conventional rodent models, being nocturnal, may not sufficiently recapitulate the bi...
Systematic single-cell analysis reveals dynamic control of transposable element activity orchestrating the endothelial-to-hematopoietic transition
The endothelial-to-hematopoietic transition (EHT) process during definitive hematopoiesis is highly conserved in vertebrates. Stage-specific expression of transposable elements (TEs) has been detected during z...
Multi-genome comparisons reveal gain-and-loss evolution of anti-Mullerian hormone receptor type 2 as a candidate master sex-determining gene in Percidae
The Percidae family comprises many fish species of major importance for aquaculture and fisheries. Based on three new chromosome-scale assemblies in Perca fluviatilis , Perca schrenkii , and Sander vitreus along wi...
- Editorial Board
- Editorial Team
- Collections
- Join the Editorial Board
- Sign up for article alerts and news from this journal
- Manuscript editing services
Annual Journal Metrics
Citation Impact 2023 Journal Impact Factor: 4.4 5-year Journal Impact Factor: 5.4 Source Normalized Impact per Paper (SNIP): 1.211 SCImago Journal Rank (SJR): 1.787 Speed 2023 Submission to first editorial decision (median days): 10 Submission to acceptance (median days): 180 Usage 2023 Downloads: 2,378,081 Altmetric mentions: 3,761
- More about our metrics
Peer Review Taxonomy
This journal is participating in a pilot of NISO/STM's Working Group on Peer Review Taxonomy, to identify and standardize definitions and terminology in peer review practices in order to make the peer review process for articles and journals more transparent. Further information on the pilot is available here .
The following summary describes the peer review process for this journal:
- Identity transparency: Single anonymized
- Reviewer interacts with: Editor
- Review information published: None.
We welcome your feedback on this Peer Review Taxonomy Pilot. Please can you take the time to complete this short survey.
Announcements
BMC Biology is recruiting new Editorial Board Members
We are looking for Editorial Board Members in all fields of biology. If you are interested in becoming an EBM please see this page .
Portable peer review
BMC Biology supports portable peer review by sharing reviews and evaluating papers based on existing reports. Learn more here .

BMC Biology is a member of the Neuroscience Peer Review Consortium.
- Follow us on Twitter
- Follow us on Facebook
BMC Biology
ISSN: 1741-7007
- General enquiries: [email protected]
- Drug resistance
- Genetic Engineering
- Microbiology
- Paleontology
- Sex & Gender
Scientists Make Living Mice’s Skin Transparent with Simple Food Dye
New research harnessed the highly absorbent dye tartrazine, used as the common food coloring Yellow No. 5, to turn tissues in living mice clear—temporarily revealing organs and vessels inside the animals
Lauren J. Young

Do Cats Really Hate Water?
Not all cats are hydrophobic
Meghan Bartels

$1 for Digital Access
Read all the stories you want.

The Earliest Known Animal Sex Chromosome is 480 Million Years Old
The octopus sex chromosome appears to have been maintained over hundreds of millions of years, making it the most ancient of such chromosomes in animals
Viviane Callier

A Dolphin That Has Been Biting People May Just Be Friendly
Dolphin ecologist Tadamichi Morisaka discusses common dolphin behaviors that could explain instances of the animals biting people in Japan
Anna Ikarashi, Nature magazine
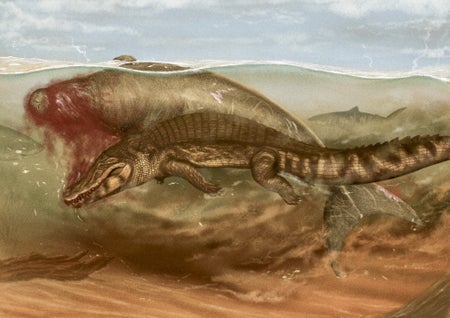
This Ancient Sea Cow Was Killed by a Croc and Eaten by a Shark
Scientists re-create the last moments of a manateelike animal that was eaten by both a crocodilian and a shark

The Pitfalls of Houseplant Collection
A curator at the New York Botanical Garden explains what we can learn about the past and the present from houseplant trends.
Rachel Feltman, Fonda Mwangi, Jeffery DelViscio

Queen’s Brian May Is a Champion for Badgers and Science
Queen guitarist Brian May has spent a decade studying the science of bovine tuberculosis, which can be carried by badgers, and has identified a new method of spread
Elizabeth Gibney, Nature magazine

Why Aging Comes in Dramatic Waves in Our 40s and 60s
A new study suggests that waves of aging-related changes occur at two distinct points in our life
Saima S. Iqbal

I Bought a Glowing Plant. It Led Me Down a Rabbit Hole
A bioluminescent petunia led me to a world of radiant mushrooms, 19th-century experiments and a modern rivalry between scientists in Russia and the Americas
Rachel Ehrenberg, Knowable Magazine

The End of the Lab Rat?
Replacing research animals with tools that better mimic human biology could improve medicine
Rachel Nuwer

What Was It Like to Be a Dinosaur?
New fossils and analytical tools provide unprecedented insights into dinosaur sensory perception
Amy M. Balanoff, Daniel T. Ksepka

Tardigrade Fossils Reveal When ‘Water Bears’ Became Indestructible
252 million years ago, tardigrades may have escaped extinction using this one weird trick
Mindy Weisberger
- Search by keyword
- Search by citation
Page 1 of 6
Endothelial adherens junctions and the actin cytoskeleton: an 'infinity net'?
A recent paper in BMC Biology reports that actin stress fibers in adjacent cultured endothelial cells are linked through adherens junctions. This organization might provide a super-cellular network that could ena...
- View Full Text
Robust and specific inhibition of microRNAs in Caenorhabditis elegans
MicroRNAs (miRNAs) are small non-coding RNAs that regulate the expression of numerous target genes. Yet, while hundreds of miRNAs have been identified, little is known about their functions. In a recent report...
Genome of a songbird unveiled
An international collaborative effort has recently uncovered the genome of the zebra finch, a songbird model that has provided unique insights into an array of biological phenomena.
The mathematics of sexual attraction
Pollen tubes follow attractants secreted by the ovules. In a recent paper in BMC Plant Biology , Stewman and colleagues have quantified the parameters of this attraction and used them to calibrate a mathematical m...
Diversity lost: are all Holarctic large mammal species just relict populations?
Population genetic analyses of Eurasian wolves published recently in BMC Evolutionary Biology suggest that a major genetic turnover took place in Eurasian wolves after the Pleistocene. These results add to the gr...
Hybridization and speciation in angiosperms: arole for pollinator shifts?
The majority of convincingly documented cases of hybridization in angiosperms has involved genetic introgression between the parental species or formation of a hybrid species with increased ploidy; however, ho...
Evolution underground: shedding light on the diversification of subterranean insects
A recent study in BMC Evolutionary Biology has reconstructed the molecular phylogeny of a large Mediterranean cave-dwelling beetle clade, revealing an ancient origin and strong geographic structuring. It seems li...
A modern circadian clock in the common angiosperm ancestor of monocots and eudicots
The circadian clock enhances fitness through temporal organization of plant gene expression, metabolism and physiology. Two recent studies, one in BMC Evolutionary Biology , demonstrate through phylogenetic analys...
Scale-eating cichlids: from hand(ed) to mouth
Two recent studies in BMC Biology and Evolution raise important questions about a textbook case of frequency-dependent selection in scale-eating cichlid fishes. They also suggest a fascinating new line of researc...
Top dogs: wolf domestication and wealth
A phylogeographic analysis of gene sequences important in determining body size in dogs, recently published in BMC Biology , traces the appearance of small body size to the Neolithic Middle East. This finding stre...
No better time to FRET: shedding light on host pathogen interactions
Understanding the spatio-temporal subversion of host cell signaling by bacterial virulence factors is key to combating infectious diseases. Following a recent study by Buntru and co-workers published in BMC Biolo...
Making progress in genetic kin recognition among vertebrates
A recent study in BMC Evolutionary Biology has shown that genetically similar individual ring-tailed lemurs are also more similar in their scent composition, suggesting a possible mechanism of kin recognition. Th...
Regeneration review reprise
There have been notable advances in the scientific understanding of regeneration within the past year alone, including two recently published in BMC Biology . Increasingly, progress in the regeneration field is be...
Acoel and platyhelminth models for stem-cell research
Acoel and platyhelminth worms are particularly attractive invertebrate models for stem-cell research because their bodies are continually renewed from large pools of somatic stem cells. Several recent studies,...
Madm (Mlf1 adapter molecule) cooperates with Bunched A to promote growth in Drosophila
The TSC-22 domain family (TSC22DF) consists of putative transcription factors harboring a DNA-binding TSC-box and an adjacent leucine zipper at their carboxyl termini. Both short and long TSC22DF isoforms are ...
Bunched and Madm: a novel growth-regulatory complex?
By combining Drosophila genetics and proteomics Gluderer et al. report in this issue of Journal of Biology the isolation of a novel growth-regulatory complex consisting of Bunched and Madm. Future study of this c...
Q&A: What can microfluidics do for stem-cell research?
Regulation of metabolism in caenorhabditis elegans longevity.
The nematode Caenorhabditis elegans is a favorite model for the study of aging. A wealth of genetic and genomic studies show that metabolic regulation is a hallmark of life-span modulation. A recent study in BMC ...
Reprogramming of the non-coding transcriptome during brain development
A recent global analysis of gene expression during the differentiation of neuronal stem cells to neurons and oligodendrocytes indicates a complex pattern of changes in the expression of both protein-coding tra...
The THO complex as a key mRNP biogenesis factor in development and cell differentiation
The THO complex is a key component in the co-transcriptional formation of messenger ribonucleoparticles that are competent to be exported from the nucleus, yet its precise function is unknown. A recent study in B...
SnoPatrol: how many snoRNA genes are there?
Small nucleolar RNAs (snoRNAs) are among the most evolutionarily ancient classes of small RNA. Two experimental screens published in BMC Genomics expand the eukaryotic snoRNA catalog, but many more snoRNAs remain...
Sometimes one just isn't enough: do vertebrates contain an H2A.Z hyper-variant?
How much functional specialization can one component histone confer on a single nucleosome? The histone variant H2A.Z seems to be an extreme example. Genome-wide distribution maps show non-random (and evolutio...
Apical polarity in three-dimensional culture systems: where to now?
Delineation of the mechanisms that establish and maintain the polarity of epithelial tissues is essential to understanding morphogenesis, tissue specificity and cancer. Three-dimensional culture assays provide...
The water flea Daphnia - a 'new' model system for ecology and evolution?
Daphnia pulex is the first crustacean to have its genome sequenced. Availability of the genome sequence will have implications for research in aquatic ecology and evolution in particular, as addressed by a series...
Top ten in Journal of Biology in 2009: stem cells, influenza, pit bulls, Darwin, and more
The bacterial pathogen listeria monocytogenes : an emerging model in prokaryotic transcriptomics.
A major challenge in bacterial pathogenesis is understanding the molecular basis of the switch from saprophytism to virulence. Following a recent whole-genome transcriptomic analysis using tiling arrays, an ar...
Forward genetics in Tribolium castaneum : opening new avenues of research in arthropod biology
A recent paper in BMC Biology reports the first large-scale insertional mutagenesis screen in a non-drosophilid insect, the red flour beetle Tribolium castaneum . This screen marks the beginning of a non-biased, '...
Mapping the protistan 'rare biosphere'
The use of cultivation-independent approaches to map microbial diversity, including recent work published in BMC Biology , has now shown that protists, like bacteria/archaea, are much more diverse than had been re...
Scribble at the crossroads
Although proteins involved in determining apical-basal cell polarity have been directly linked to tumorigenesis, their precise roles in this process remain unclear. A recent report in BMC Biology clarifies the si...
Q&A: Quantitative approaches to planar polarity and tissue organization
Gene regulation, evolvability and the limits of genomics, the transcriptome of human monocyte subsets begins to emerge.
Human monocytes can be divided into subsets according to their expression or lack of the cell-surface antigen CD16. In papers published recently in the Journal of Proteome Research and in BMC Genomics , two groups...
Chromatin 'programming' by sequence - is there more to the nucleosome code than %GC?
The role of genomic sequence in directing the packaging of eukaryotic genomes into chromatin has been the subject of considerable recent debate. A new paper from Tillo and Hughes shows that the intrinsic therm...
Fishing for the signals that pattern the face
Zebrafish are a powerful system for studying the early embryonic events that form the skull and face, as a model for human craniofacial birth defects such as cleft palate. Signaling pathways that pattern the p...
Coordinated gene expression by post-transcriptional regulons in African trypanosomes
The regulation of gene expression in trypanosomes is unique. In the absence of transcriptional control at the level of initiation, a subset of Trypanosoma brucei genes form post-transcriptional regulons in which ...
Promoter architecture and the evolvability of gene expression
Evolutionary changes in gene expression are a main driver of phenotypic evolution. In yeast, genes that have rapidly diverged in expression are associated with particular promoter features, including the prese...
Adaptations of proteins to cellular and subcellular pH
Bioinformatics-based searches for correlations between subcellular localization and pI or charge distribution of proteins have failed to detect meaningful correlations. Recent work published in BMC Biology finds ...
TBP2 is a general transcription factor specialized for female germ cells
The complexity of the core promoter transcription machinery has emerged as an additional level of transcription regulation that is used during vertebrate development. Recent studies, including one published in BM...
Generalized immune activation as a direct result of activated CD4 + T cell killing
In addition to progressive CD4 + T cell immune deficiency, HIV infection is characterized by generalized immune activation, thought to arise from increased microbial exposure resulting from diminishing immunity.
Life and death as a T lymphocyte: from immune protection to HIV pathogenesis
Detailed analysis of T cell dynamics in humans is challenging and mouse models can be important tools for characterizing T cell dynamic processes. In a paper just published in Journal of Biology , Marques et al . s...
What we still don't know about AIDS
The gene complement of the ancestral bilaterian - was urbilateria a monster.
Expressed sequence tag analyses of the annelid Pomatoceros lamarckii , recently published in BMC Evolutionary Biology , are consistent with less extensive gene loss in the Lophotrochozoa than in the Ecdysozoa, but ...
The nature of cell-cycle checkpoints: facts and fallacies
The concept of checkpoint controls revolutionized our understanding of the cell cycle. Here we revisit the defining features of checkpoints and argue that failure to properly appreciate the concept is leading ...
An expanded evolutionary role for flower symmetry genes
CYCLOIDEA (CYC) -like TCP genes are critical for flower developmental patterning. Exciting recent breakthroughs, including a study by Song et al. published in BMC Evolutionary Biology , demonstrate that CYC -like ge...
Mechanisms of ubiquitin transfer by the anaphase-promoting complex
The anaphase-promoting complex (APC) is a ubiquitin-protein ligase required for the completion of mitosis in all eukaryotes. Recent mechanistic studies reveal how this remarkable enzyme combines specificity in...
Targeting TNF-α for cancer therapy
As the tumor vasculature is a key element of the tumor stroma, angiogenesis is the target of many cancer therapies. Recent work published in BMC Cell Biology describes a fusion protein that combines a peptide pre...
TEs or not TEs? That is the evolutionary question
Transposable elements (TEs) have contributed a wide range of functional sequences to their host genomes. A recent paper in BMC Molecular Biology discusses the creation of new transcripts by transposable element i...
Molecular machines or pleiomorphic ensembles: signaling complexes revisited
Signaling complexes typically consist of highly dynamic molecular ensembles that are challenging to study and to describe accurately. Conventional mechanical descriptions misrepresent this reality and can be a...
Ockham's broom: A new series
Adaptation by introgression.
Both selective and random processes can affect the outcome of natural hybridization. A recent analysis in BMC Evolutionary Biology of natural hybridization between an introduced and a native salamander reveals th...
Journal of Biology
ISSN: 1475-4924
- Search Menu
- Sign in through your institution
- Advance Articles
- Virtual Issues
- High-Impact Research Collection
- Celebrate 40 years of MBE
- Perspectives
- Discoveries
- Cover Archive
- Brief Communications
- Submission site
- Author guidelines
- Open access
- Self-archiving policy
- Reasons to submit
- About Molecular Biology and Evolution
- About the Society for Molecular Biology and Evolution
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic

Article Contents
Introduction, acknowledgments, data availability, epigenetics research in evolutionary biology: perspectives on timescales and mechanisms.
- Article contents
- Figures & tables
- Supplementary Data
Soojin V Yi, Epigenetics Research in Evolutionary Biology: Perspectives on Timescales and Mechanisms, Molecular Biology and Evolution , Volume 41, Issue 9, September 2024, msae170, https://doi.org/10.1093/molbev/msae170
- Permissions Icon Permissions
Epigenetics research in evolutionary biology encompasses a variety of research areas, from regulation of gene expression to inheritance of environmentally mediated phenotypes. Such divergent research foci can occasionally render the umbrella term “epigenetics” ambiguous. Here I discuss several areas of contemporary epigenetics research in the context of evolutionary biology, aiming to provide balanced views across timescales and molecular mechanisms. The importance of epigenetics in development is now being assessed in many nonmodel species. These studies not only confirm the importance of epigenetic marks in developmental processes, but also highlight the significant diversity in epigenetic regulatory mechanisms across taxa. Further, these comparative epigenomic studies have begun to show promise toward enhancing our understanding of how regulatory programs evolve. A key property of epigenetic marks is that they can be inherited along mitotic cell lineages, and epigenetic differences that occur during early development can have lasting consequences on the organismal phenotypes. Thus, epigenetic marks may play roles in short-term (within an organism's lifetime or to the next generation) adaptation and phenotypic plasticity. However, the extent to which observed epigenetic variation occurs independently of genetic influences remains uncertain, due to the widespread impact of genetics on epigenetic variation and the limited availability of comprehensive (epi)genomic resources from most species. While epigenetic marks can be inherited independently of genetic sequences in some species, there is little evidence that such “transgenerational inheritance” is a general phenomenon. Rather, molecular mechanisms of epigenetic inheritance are highly variable between species.
This perspective is part of a series of articles celebrating 40 years since Molecular Biology and Evolution was founded. It is accompanied by virtual issues on this topic published by Genome Biology and Evolution and Molecular Biology and Evolution, which can be found at our 40th anniversary website .
There has been much interest in exploring epigenetics in the study of evolutionary biology, as evidenced by numerous articles in the pages of Molecular Biology and Evolution and Genome Biology and Evolution. I became interested in epigenetics and its impacts on genome evolution nearly two decades ago, when researchers began discovering DNA methylation in many taxa where it was previously thought to be absent (e.g. [ Wang, et al. 2006 ; Grbic, et al. 2011 ; Gao, et al. 2012 ]). Since then, a notable “paradigm shift” has occurred contrasting with the earlier view that DNA methylation had limited impact across taxa, bringing considerable recognition to the widespread presence of DNA methylation throughout the tree of life.
The current consensus is that DNA methylation of gene bodies (“gene body DNA methylation”) was ancestrally present and that it has undergone lineage-specific changes, including losses in lineages containing the iconic laboratory model species Drosophila melanogaster and Caenorhabditis elegans (e.g. [ Werren, et al. 2010 ; Yi 2012 ; Zhong 2016 ; de Mendoza, et al. 2020 ]). A notable event in animal evolution was the emergence of genome-wide DNA methylation (not just gene body DNA methylation) in the early stages of vertebrate evolution ( Tweedie, et al. 1997 ; Suzuki and Bird 2008 ; Keller, et al. 2015 ; Angeloni, et al. 2024 ), although there are still some human genes (and likely other vertebrate genes) that are devoid of DNA methylation altogether ( Mendizabal, et al. 2017 ). Remarkably, it is becoming increasingly clear that genomic patterns of DNA methylation can change rapidly, even between closely related species ( Bewick, et al. 2016 , 2017 ; Schmitz, et al. 2019 ; Sarkies 2022 ; Sadler 2023 ).
Conservation and divergence of histone modifications during evolution are less well understood than those of DNA methylation. Nevertheless, it is well established that several components of histone modifications and nucleosomes predated the emergence of eukaryotes ( Sandman and Reeve 2006 ; Ammar, et al. 2012 ; Erives 2017 ; Talbert, et al. 2019 ; Grau-Bové, et al. 2022 ). On the other hand, histone modifications of regulatory regions are shown to evolve rapidly between species, especially those associated with intergenic regulatory elements such as enhancers ( Villar, et al. 2015 ; Garcia-Pérez, et al. 2021 ).
Our understanding of the epigenome evolution has significantly improved over the past two decades since I first entered this field. We now know that the epigenomes are phylogenetically widespread and that epigenetic modifications can evolve rapidly. These recent paradigm shifts have introduced even more intriguing and unresolved questions about the associated consequences of epigenomic evolution on the genome. The importance of epigenetic changes as powerful mediators of phenotypes during evolution is becoming increasingly recognized, and comparative epigenomic studies are providing promising tools to understand genome regulation and annotation. The study of epigenetically mediated phenotypes has the potential to advance our understanding of molecular mechanisms of adaptation. Here I discuss these contemporary areas of research while pointing out important considerations that can help resolve several misunderstandings regarding the term epigenetics. Most of my experience with epigenetics comes from studies of DNA methylation, which is arguably the most phylogenetically broadly studied epigenetic mark. Consequently, many examples are from DNA methylation, although I have included some examples of other epigenetic marks.
The Focus of Epigenetic Studies is Highly Divergent in Different Contexts
The term “ epigenetics ” is used broadly, and sometimes ambiguously, across divergent realms of scientific literature. At the molecular level, epigenetics is the study of “epigenetic marks” that chemically modify biological molecules including genomic DNA, RNA, and proteins. Widely studied epigenetic marks include methylation of DNA (DNA methylation) and chemical modifications of histone subunits of nucleosomes. Small RNA molecules that influence genome integrity, such as piRNAs, are also considered epigenetic marks ( Zhang, et al. 2020 ). Together, these marks affect how different sections of the genome are packaged, altering the three-dimensional structures and molecular accessibility of the genomic regions (e.g. [ Cavalli and Misteli 2013 ; Allis and Jenuwein 2016 ]) within each and every nucleus of cells within an organism.
There is an especially prominent divide in the use of the term epigenetics between molecular genetic literature and ecological literature ( Deans and Maggert 2015 ). Broadly speaking, the former focuses on molecular mechanisms of expression changes, while the latter tends to concern environmentally mediated phenotypes ( Deans and Maggert 2015 ). These two areas also deal with divergent timescales, the former within an organism's lifetime, and the latter within longer timescales that can span generations. In addition, due to the limited genomic and molecular resources of nonmodel species, the latter often uses organismal phenotypes as readouts.
From my personal experience, some evolutionary biologists who are not necessarily involved in epigenetics research tend to be more familiar with the term “epigenetics” as it is commonly used in ecological literature (associated with environmentally mediated phenotypes). In reality, many evolutionary epigenetic studies focus on the molecular mechanisms of expression changes and genome regulation between species, and thus use the term “epigenetics” in the manner more typical for molecular genetic literature. The misunderstandings and complexities that arise due to the ambiguity of the term epigenetics have been discussed elsewhere (e.g. [ Deans and Maggert 2015 ; Richards, et al. 2017 ]). Scientists may be thinking about entirely different phenomena if the term epigenetics is used without clarifying details. In principle, it may be possible to bridge these divergent scales through molecular mechanisms that explain how epigenetic marks influence cellular phenotypes (referred to as “epigenetic mechanisms” in this article) and how these marks are inherited across cell generations. However, it is becoming clear that epigenetic mechanisms may be highly variable between different taxa, and sometimes even within the same species depending on the genetic background and/or molecular methods used to study them, as I will demonstrate below. Consequently, extrapolating the impacts of epigenetics across different timescales and between taxa can be misleading.
Epigenetic Marks are key Regulators of Development, but the Specific Outcomes are Dependent on Genetic Background
Epigenetic marks are known for their crucial roles in developmental processes. Importantly, DNA methylation marks, and some histone modifications, are transmitted through mitotic cell divisions ( Probst, et al. 2009 ; Alabert, et al. 2017 ; Brickner 2023 ). Consequently, these epigenetic marks are largely conserved in descendant cell populations, or “cell lineages,” although spontaneous mutations of epigenetic marks, referred to as “epimutations,” are also known to arise (see [ Bogan and Yi 2024 ] for discussion on this topic). As a result, epigenetic marks laid during early development will have long-lasting functional consequences during the lifetime of an organism via impacts on long-lived cell lineages and their descendant cell populations. This concept is well displayed in the so-called “epigenetic landscape” figure by Waddington ( Waddington 1957 ), where the “epigenetic state” of different cell populations and their descendants is depicted as a ball following through different downward paths, analogous to different cell lineage fates during organismal development (See Fig. 1 for the author's modification of the epigenetic landscape). More broadly, variation in epigenetic marks during any point in development can influence many subsequent cell generations.

A modified version of the “epigenetic landscape” in Waddington (1957) . Stem cell populations (near the top of the landscape) experiencing different epigenetic modifications follow different “paths” toward more differentiated states, resulting in populations of cells that harbor distinct epigenetic marks from other cell populations. Each population of differentiated cells is specialized for distinct cellular niches. Figure is modified from Yi (2017) .
The critical importance of epigenetic mechanisms in development is supported by a myriad of studies, yet this research has also revealed substantial differences between taxa. I will demonstrate this point using studies that examined the consequences of modulating key DNA methylation enzymes, named “DNA methyltransferases ( dnmt s)” ( Goll and Bestor 2005 ; Lyko 2018 ). These are one of the most conserved proteins in the tree of life. Starting with mammalian studies, mutations of dnmt s result in lethality in mice embryos ( Li, et al. 1992 ; Okano, et al. 1999 ). Mutations in human and mouse dnmt s in somatic cells are also generally lethal (e.g. [ Trowbridge, et al. 2009 ]). Deletion of dnmt1 in human embryonic stem cells results in cell death ( Liao, et al. 2015 ). Mutations in other epigenetic regulators cause developmental disorders and cancers, and phenotypic alterations of multiple organs ( Feinberg 2007 ).
Similar studies in nonmodel animal species are much rarer compared to those in mammals. While they generally support the importance of epigenetic marks in development, the degree and severity of the consequences of dnmt manipulation vary considerably across different studies, depending on the methods used and the developmental stages when the manipulations were performed. For example, knockdown of a dnmt3 homolog in newly hatched honey bee larvae changed the course of development between queen- and worker-like phenotypes ( Kucharski, et al. 2008 ). Knockdown of a dnmt1 homolog using parental RNAi in a parasitic wasp Nasonia vitripennis resulted in developmental failure and embryonic lethality ( Arsala, et al. 2022 ). In the milkweed bug Oncopeltus fasciatus , knockdown of a dnmt1 homolog in embryos negatively affected testis development and resulted in the reduction of genome-wide DNA methylation ( Bewick, et al. 2019 ; Washington, et al. 2021 ). In the clonal raider ant Ooceraea biroi, mutating dnmt1 homolog using CRISPR/Cas9 in eggs resulted in sterile individuals with a significantly reduced lifespan ( Ivasyk, et al. 2023 ). Knockout of dnmt3a isoform using CRISPR/Cas9 in zebrafish embryos resulted in organismal deaths in a temperature-dependent manner ( Loughland, et al. 2021 ). These results support that epigenetic marks are important regulators of development and cell lineage differentiation in the studied animals. However, apart from changes of genome-wide DNA methylation, molecular consequences of dnmt changes on gene expression are not consistent across animal studies, and some authors suggest DNA methylation may play a role in reproduction via (an) unrelated mechanism(s) to gene expression (e.g. [ Amukamara, et al. 2020 ; Washington, et al. 2021 ; Ivasyk, et al. 2023 ]). Therefore, DNMT1 orthologs may have important regulatory functions aside from DNA methylation. One confounding factor in these studies may be cell heterogeneity. As we learn that epigenetic marks in different cell types are highly distinctive ( Kundaje, et al. 2015 ; Cusanovich, et al. 2018 ; Mendizabal, et al. 2019 ), assessing epigenetic changes and their consequences in samples consisting of heterogeneous cell types could “mask” the true causal effects due to the lack of necessary resolution.
Studies in plants reveal yet another degree of divergence. In rice and maize, loss-of-function of single DNA methyltransferase results in lethality ( Hu, et al. 2014 ; Fu, et al. 2018 ). On the other hand, in Arabidopsis thaliana , loss-of-function mutants of one or more DNA methyltransferases, including those orthologous to animal dnmt1 , exhibit relatively minor phenotypic impacts ( Kankel, et al. 2003 ; Stroud, et al. 2014 ), although more severe effects were also observed ( Mathieu, et al. 2007 ). Even A. thaliana individuals with four out of the five DNA methyltransferase deleted were viable and exhibited weak phenotypic differences such as curled leaves and reduced rosette size ( Henderson and Jacobsen 2008 ; Stroud, et al. 2014 ). Two recent studies reported mutants where all five currently known DNA methyltransferases were deleted from A. thaliana ( He, et al. 2022 ; Liang, et al. 2022 ). In one study, the quintuple mutants were still viable, although they exhibited serious developmental delays, including small size and the inability to undergo floral transition ( He, et al. 2022 ). However, in another study, the resulting quintuples were nonviable ( Liang, et al. 2022 ). These two studies utilized different genetic backgrounds to generate the quintuples. These examples highlight that the consequences of experimental manipulations of key epigenetic enzymes are highly divergent in different taxa, and even in the same species depending on specific genetic backgrounds and molecular methods used, a theme that is repeatedly observed in other areas of research as I discuss below.
Promises and Challenges of Comparative Epigenetic Studies
Given its importance in development and cell lineage differentiation, and the rapid evolutionary changes of epigenetic marks across the tree of life, comparative studies of epigenetic marks may advance our understanding of the evolution of development, phenotypic divergence, and genome evolution. More broadly, epigenetic mechanisms make attractive candidate causative agents of regulatory evolution, a prime topic of interest for many molecular evolutionists.
Consequently, there is a growing interest in the comparative analysis of epigenetic marks. An important concern in designing comparative epigenomic studies is to select specific tissues/cell types, since epigenetic marks are differentiated between tissues and cell types ( Villar, et al. 2015 ; Jeong, et al. 2021 ; Singh, et al. 2021 ; Caglayan, et al. 2023 ; Klughammer, et al. 2023 ). Most of the current literature is on the evolution of DNA methylation. While some earlier studies used microarrays, bisulfite sequencing is widely used to generate species-specific DNA methylation maps (e.g. [ Molaro, et al. 2011 ; Zeng, et al. 2012 ; Glastad, et al. 2014 ; Mendizabal, et al. 2016 ; Jeong, et al. 2021 ; Al Adhami, et al. 2022 ; Hu, et al. 2023 ; Chen, et al. 2024 ]). In addition, it is important to consider bisulfite conversion rates, and the presence of C to T SNPs that affect mapping. In the best possible practice, bisulfite conversion rates should be independently estimated in different experiments and C to T SNPs should be masked based on genome sequence data (e.g. [ Jeong et al 2021 , Wu et al. 2000 ]).
While it is relatively straightforward to identify DNA methylation differences between species, deciphering whether and how the observed differences contribute to functional differences is challenging, given our incomplete understanding of molecular mechanisms of epigenetics. In terms of DNA methylation literature, one of the most widely applied ideas is the cis-regulatory effect of DNA methylation in silencing gene expression ( Schübeler 2015 ). However, this relationship is applicable to only a subset of promoters, most likely those that use CpG rich sequences, or CpG islands. For other promoters, there is often no relationship or even negative relationship between promoter methylation and gene expression (e.g. [ Morgan et al. 2024 ]). The relationship between DNA methylation and expression in other genomic contexts is even less clear. For example, the role of gene body methylation is still highly debated (e.g. [ Jones 2012 ; Huh, et al. 2013 ; Hunt et al. 2013 ; Takuno and Gaut 2013 ; Bewick and Schmitz 2017 ; Zilberman 2017 ; Seymour and Gaut 2020 ; Wu, et al. 2022 ]). Candidate gene studies focusing on specific genes of interest show some success in identifying functional epigenetic differences. For example, a study of ADRA2C in primate brains identified previously uncharacterized cis-regulatory regions that could drive gene expression in reporter assays ( Lee, et al. 2018 ). This region harbored divergent epigenetic signatures between primate brains. Specifically, hypermethylation of the newly identified cis-regulatory region in the human and chimpanzee brains coincided with the increase of expression, associated with fight-or-flight response ( Lee, et al. 2018 ). In another study, human-specific hypomethylation of the 5′ UTR region of a CENPJ, a gene associated with brain size increase, was discovered in a comparative study of several human and nonhuman primates ( Shi, et al. 2014 ).
Notwithstanding the difficulties associated with inferring functional mechanisms, comparative epigenetic data themselves can be useful to identify functional regions, especially from noncoding regions of the genome. For example, analysis of DNA methylation across multiple tissues in primates demonstrated that differentially methylated regions (DMRs) between tissues tended to be conserved between primate species, and genes near those evolutionary DMRs tended to encode tissue-relevant functional annotations ( Blake, et al. 2020 ). A recent study of muscle and liver tissues across 13 mammals reported that methylation changes of promoter regions coincided with genes that encode species-specific biological traits ( Hu, et al. 2023 ). Furthermore, comparative epigenetic studies can even provide insights into molecular mechanisms of epigenetic marks. For example, DNA methylation was thought to play important roles in the regulation of X chromosome inactivation by methylating the inactive X chromosome. Comparative studies reveal that the methylation of the inactive X chromosomes is not conserved in mammals. Instead, the loss of DNA methylation of the inactive X chromosomes is widely observed ( Weber, et al. 2005 ; Hellman and Chess 2007 ; Singh, et al. 2021 ; Morgan, et al. 2024 ). Therefore, comparative epigenetic studies hold promise to identify potential regulatory regions, and to provide useful information in understanding the mechanisms of how epigenetic marks influence genome regulation. While these efforts have mostly focused on vertebrates, there is potential to use epigenetic information to identify regulatory positions in invertebrate genomes (e.g. [ Jeong et al. 2018 ]). It should be noted that epigenomic differences observed between species by no means imply that they are independent of genetic differences. Rather, some, if not the majority of the epigenetic differences found between species, may have genetic origins, as there is strong evidence that sequence contexts are important drivers of epigenome patterns, at least for DNA methylation (e.g. [ Lienert, et al. 2011 ; Krebs, et al. 2014 ; Long, et al. 2016 ]).
Potential Link between Epigenetics and Phenotypic Plasticity Needs to Consider Genetic Effects and Divergent Molecular Mechanisms
Since epigenetic changes at one point in a lifetime of an organism can affect the target cell populations and its descendant cells, epigenetic variation can theoretically produce phenotypic impacts at various timescales within an organism's lifespan. For example, epigenetic variation at an early developmental time point could lead to different phenotypic trajectories of adults. One such possibility was raised regarding the polyphenism in honey bees. When a DNA methylation enzyme dnmt3 was knocked out using RNAi, larval honey bees preferentially developed into queen-like, rather than worker-like, phenotypes ( Kucharski, et al. 2008 ). These results supported the idea that variation of DNA methylation leads to different phenotypes of bees. Another classic example is the “ Agouti viable yellow ( Avy )” mouse, where genetically identical mouse individuals exhibit different coat colors. In this system, the coat color difference was caused by variable DNA methylation of a transposable element ∼100 kb upstream of the Agouti coat color locus ( Duhl, et al. 1994 ; Morgan, et al. 1999 ). Additionally, the coat color variation was influenced by maternal dietary supplementation of methyl donors and co-factors ( Wolff, et al. 1998 ; Cropley, et al. 2006 ). These studies emboldened the Avy mouse system as an example of an epigenetic phenotypic trait that could be mediated by environmental effects, in this case diet ( Bertozzi and Ferguson-Smith 2020 ). There are numerous studies that have explored epigenetic variation associated with phenotypic plasticity in other contexts (e.g. [ Roberts and Gavery 2012 ; Zhang, et al. 2013 ; Duncan, et al. 2014 ; Richards, et al. 2017 ; Springer and Schmitz 2017 ; Loughland, et al. 2021 ]). There are abundant examples where parental experience is inherited to their immediate offsprings, likely in the form of epigenetic marks, and that such epigenetic inheritance may confer selective advantages ( Heard and Martienssen 2014 ; Rechavi and Lev 2017 ; Fitz-James and Cavalli 2022 ).
Some authors thus hypothesized that environmentally induced epigenetic variation, independent of genetic variation, could produce adaptive phenotypes ( Bossdorf, et al. 2008 ; Jablonka and Raz 2009 ). However, there are several challenges to connecting the environment to epigenetic variation, and subsequently epigenetic variation to adaptation. These include the confounding effects of genetics, methodological difficulties, and uncertain molecular mechanisms. A key issue that is widespread in literature is an inadequate consideration of genetic effects. It is becoming clear that epigenetic variation is tightly linked to genetic variation in a variety of taxa (e.g. [ Wang, et al. 2016 ; Petronis 2010 ; Eitchen et al 2013 ; Schmitz, et al. 2013 ; Seymour and Becker 2017 ; Villicaña and Bell 2021 ; Hämälä, et al. 2022 ; Sepers, et al. 2023 ]). Therefore, any study of epigenetic variation between individuals or populations should take into account the underlying genetic variation as a source of epigenetic variation. Unfortunately, this is extremely difficult to do, because genetic drivers of epigenetic variation are pervasive in the genome, and many species lack detailed genetic data to account for the genetic effects. For example, DNA methylation studies of human populations, where large cohorts and abundant molecular resources are available, observe that there are numerous methylation quantitative trait loci (meQTLs) that affect DNA methylation. These meQTLs can function both in cis - and trans -, and are found across the whole genome ( Villicaña and Bell 2021 ). Incorporating meQTLs into epigenomic studies reveal that it is notoriously difficult to separate the effect of underlying genetics in the study of epigenetic variation ( Do, et al. 2017 ; Gao, et al. 2017 ). For example, in a study of genome-wide epigenetic variation in human immune cells including neutrophils, monocytes and T-cells, more than 50% of epigenetic variation was attributed to genetic effects ( Chen, et al. 2016 ). The well-known example of the Avy mouse mentioned above also shows genetic background effects, exhibiting less prominent effects in different maternal backgrounds ( Wolff 1978 ). Therefore, we still lack a thorough understanding of the amount of truly “epigenetic” variation that exists independently of genetic variation in nature.
Even if genetic effects could be successfully accounted for, it is challenging to connect an observed epigenetic variant to its adaptation to the environment. At minimum, we should observe reproducible epigenetic patterns independent of genetic variation and the specific epigenetic phenotype of interest having higher fitness than others, both of which have proven difficult to address in many instances. Another difficulty is identifying the molecular loci whose epigenetic difference causes functional differences. For example, in the example of the Avy mouse, dietary supplementation (environmental effect) was shown to influence the color phenotype ( Cooney, et al. 2002 ; Waterland and Jirtle 2003 ; Cropley, et al. 2006 ). This was taken as a support for the hypothesis that dietary supplementation of methyl donors led to an increase of DNA methylation of the ‘causative’ transposable element. However, no methylation difference was observed in the said candidate transposable element in mice with or without dietary supplementation ( Cropley, et al. 2010 ). In the case of the aforementioned honey bee example, it was pointed out that the RNAi was performed after the phenotypic fates of larvae were already determined ( Duncan, et al. 2022 ), indicating that the manipulation of DNA methylation was not the only effect measured by the phenotypes.
Therefore, even though there are examples of environmentally induced phenotypes associated with epigenetic differences, whether they are truly independent of genetic effects has not been exhaustively evaluated in many studies, in large part due to lack of genetic resources to enable such analysis in nonmodel species. When genetic resources are available (as in the example of mice), in-depth studies revealed that phenotypic effects were sometimes inconsistent in different genetic backgrounds, and the molecular mechanisms of environmental effects were not necessarily replicated. Furthermore, long-term adaptive consequences of such changes and the underlying molecular mechanisms of such processes remain elusive. Interested readers in this topic should refer to extensive reviews in literature (e.g. [ Venney, et al. 2023 ; Bogan and Yi 2024 ]). Nevertheless, ecological epigenetic studies have a great potential to reveal causative mechanisms of gene expression changes and genome regulation ( Richards, et al. 2017 ).
Epigenetic Inheritance is not Universal and its Adaptive Significance is Unresolved
In some species, epigenetic marks can be inherited independently of genetic sequences. Does this mean that epigenetic marks themselves can be considered an additional component of inheritance? If so, can they provide the raw materials of adaptive evolution? These questions have sparked substantial interest in recent years. Researchers in these areas often focus on environmentally induced epigenetic variation. I will discuss a few key questions in this regard.
First, is there sufficient epigenetic variation in nature? Most certainly (e.g. [ Carja, et al. 2017 ; Noshay and Springer 2021 ]). Variation of epigenetic marks, or epialleles (defined as alleles harboring different epigenetic marks), can arise via several mechanisms, some due to genetic variation, some independent of genetics (see [ Bogan and Yi 2024 ] for more details). For example, in many species, epigenetic variation is known to arise during the normal process of aging, even for young, reproductive individuals (e.g. [ Pal and Tyler 2016 ; Lu et al. 2023 ; Brink et al. 2024 ]). Stochastic mutations of epigenetic marks, or “epimutations,” also occur. The most well characterized epimutations have been from the studies of A. thaliana and C. elegans . In A. thaliana , DNA methylation studies of carefully curated recombinant inbred lines revealed extensive epimutations ( Becker, et al. 2011 ; Schmitz, et al. 2013 ) and much higher rates of epimutations than those of point mutations ( van der Graaf, et al. 2015 ). In C. elegans , small RNAs in mutation accumulation lines indicated stochastic variations of small RNA abundance, at much higher rates than point mutations ( Beltran, et al. 2020 ). It is worth noting that data from other taxa is currently lacking so we should refrain from extending these observations to broader contexts. In addition, these two well-known examples originate from two highly divergent molecular pathways, once again cautioning against generalizing these observations in a broad category of “epimutations.” Second, do environmental changes cause epimutations? There are conflicting reports in the literature. Some studies report an increase of epimutations under environmental stress (e.g. [ Jiang, et al. 2014 ]), while others report epigenomes that are largely stable (e.g. [ Hämälä, et al. 2022 ]).
Third, can the epialleles be transmitted through generations? Or, do epigenetic marks experience the so-called “transgenerational inheritance”? This is a question that many researchers are interested in. It should be noted that many studies propose transgenerational inheritance based on phenotypic observations, but the molecular demonstration of the transgenerational inheritance of epigenetic marks is limited, and current evidence is mostly from A. thaliana and C. elegans . In A. thaliana , epialleles were shown to be transmitted through several generations ( Johannes, et al. 2009 ). The transmission of epialleles to germline may be facilitated by their reproductive biology where the germline cells arise from somatic cells. In C. elegans , transgenerational inheritance of small RNAs across multiple generations has been observed ( Ashe, et al. 2012 ; Rechavi, et al. 2014 ). The inheritance of small RNAs in C. elegans is orchestrated by movement of small RNAs between cells including oocytes, as well as RNA-amplification mechanisms ( Rechavi and Lev 2017 ).
In mammals, the presence and/or mechanisms of transgenerational inheritance are still inconclusive. It is important to discuss further details of mammalian studies. In the example of the Avy mouse, supplementing mother’s diet with methyl donors influenced offspring coat colors. However, this effect was not transmitted for more than two generations, therefore disputing the existence of a “transgenerational” effect ( Daxinger and Whitelaw 2012 ). Rather, this could be viewed as an example of an “intergenerational” effect where epigenetic variation in parents impacts offspring development, of which there are a number of examples ( Heard and Martienssen 2014 ). In another widely cited example, exposing pregnant female rats to a fungicide vinclozolin impacted male reproductive traits in progenies including F3 and F4 generations ( Anway, et al. 2005 ). However, this observation was not replicated in other genetic backgrounds ( Schneider, et al. 2008 , 2013 ; Inawaka, et al. 2009 ). It was proposed that using inbred lines may have obscured epigenetic inheritance ( Hanson and Skinner 2016 ), but the underlying logic is unclear. It is possible that the outbred lines harbored genetic polymorphism that enabled the observed phenotypes. To identify the actual epigenetic marks that underlie the observed transgenerational inheritance, sperm methylomes of these mice were compared, identifying many DMRs between the control and the vinclozolin-treated lines. However, there was no DMR that was consistently found in the F1 and F3 generations, ruling out a consistent differential methylation of specific loci as an underlying mechanism ( Beck, et al. 2017 ).
A recent study took a different approach and started with the molecular marks by generating DNA methylation-edited mice ( Takahashi, et al. 2023 ). The authors induced DNA methylation of a specific CpG island near the Ankryn repeat domain 26 ( Ankrd26 ) locus in mice ( Takahashi et al. 2023 ) by inserting a CpG-free insert near that region of embryonic stem cells. Note that this experimental scheme relied on the cis-effect of genomic sequence influencing DNA methylation. The authors then used sophisticated gene editing tools to excise out the CpG-free insert, theoretically reverting the cells to the previous genetic background while maintaining the gain of DNA methylation of the target CpG island. They then injected the resulting cells to 8-cell embryos of another strain of mouse, and demonstrated that DNA methylation of this CpG island was maintained for three generations in some, but not in all, cells. While this approach is exciting in that it directly followed the transmission of an epigenetic mark, it is still unclear if the induced cells were entirely of the same genetic background as those prior to the induction of DNA methylation. Indeed, DNA methylation of the induced cells were altered not just at the target CpG island, but over two dozen of other genomic regions, including some where DNA methylation was changed by 91% ( Takahashi et al. 2023 ). It is possible that the gene editing tools they used modified other genomic regions that influence DNA methylation. Even though the authors confirmed the absence of point mutations within 1 million base pairs of the target site, trans -meQTLs are known to influence DNA methylation of distant genomic regions ( Villicaña and Bell 2021 ). In fact, the observed transgenerational inheritance was specific to the induction-excision method they used, and was not observed when a different method, dCas9-DNA methyltransferase system, was used to modify DNA methylation ( Takahashi, et al. 2023 ).
In summary, although many studies strive to investigate the presence and/or mechanisms of transgenerational inheritance in mammals, the exact molecular changes prove elusive. So far, well-known examples have been somewhat restrictive to either the specific genetic background or the molecular method used. As a result, we must be wary of generalizing these observations. Furthermore, if these patterns manifest so temperamentally in well-controlled laboratory conditions, it is hard to expect stable long-term epigenetic inheritance in nature.
Fourth, do epigenetic variations confer adaptive potential? This question is difficult to address in nonmodel species due to the tight association between genetic and epigenetic variation, and the relative lack of (epi)genomic resources. Studies using A. thaliana reported potential adaptive changes of DNA methylation and transgenerational inheritance (e.g. [ Bossdorf, et al. 2010 ; Latzel, et al. 2013 ]) but recent studies utilizing whole genome methylomes report more nuanced or negative results (e.g. [ Ganguly, et al. 2017 ; Van Dooren, et al. 2020 ]). Here it is useful to consider evolutionary consequences if epigenetic variations were indeed functional (which would be implied if they were to be adaptive). Studies of genetic mutations have demonstrated that the majority of functional (i.e. nonneutral) mutations are deleterious, rather than advantageous. Given that rates of epigenetic mutations are much higher than those of genetic mutations, the majority of epigenetic variation in nature is likely to be neutral with little functional consequences, simply because of the high genetic load of frequent functional mutations ( Charlesworth, et al. 2017 ). Population genetic analyses of epigenetic variation may be critically informative to resolve the selective potential of epigenetic variation. Such studies so far have reported extremely weak selective effects of DNA methylation variation in A. thaliana ( Vidalis, et al. 2016 ; Muyle, et al. 2021 ). These observations suggest that much like genetic mutations, a large portion of epigenetic variation may be neutral with occasional rare advantageous epigenetic mutations occurring in specific genetic background and conditions, useful in short-term adaptation.
Concluding Remarks
I discussed several areas of epigenetics research in the context of evolutionary biology. The topics I discussed in this article highlight several key considerations: first, we must strive to adequately consider genetics when analyzing epigenetic variation. This is not only useful to infer the extent of “true” epigenetic variation but also to improve our understanding of genome regulation and adaptation via epigenetic mechanisms. Second, we cannot generalize findings in one experimental system to broader contexts, as there is significant variation within species, not to mention between species. I discussed that even DNA methylation, which is a highly conserved and extensively studied epigenetic mechanism of development, can have different functional consequences in different taxa or even within the same species. The prevalence and the molecular mechanisms of transgenerational inheritance are highly divergent between different species, due to differences in cellular and reproductive biology. Third, a significant portion of the literature relies on phenotypic readouts to infer underlying epigenetic inheritance, but when the detailed molecular mechanisms of epigenetic marks are investigated, they are often much more complex and obscure. This is partly due to the difficulty of inferring the molecular basis of quantitative phenotypic traits, but could also indicate strong genome-epigenome interactions that depend on genetic background and specific conditions.
Needless to say, this article is not meant to be a comprehensive discussion of epigenetics in evolutionary biology and there are many other exciting areas of research. One such area is the study of genome-epigenome interactions. Epigenetic marks such as DNA methylation and histone modification can directly influence genome sequence evolution (e.g. [ Makova and Hardison 2015 ; Yi and Goodisman 2021 ]). Conversely, genome sequences can affect epigenome configuration. This area of research has a great potential to enhance our understanding of the tempo and dynamics of genome evolution and help functional annotation of genome sequences, especially of noncoding regions. Transposable elements are key components of epigenetic inheritance, and we cannot understand the evolution of epigenomes without understanding the evolution of transposable elements. Transposable elements also influence mutation rates and gene expression of the genomic neighborhoods where they reside (e.g. [ Hollister and Gaut 2009 ; Choi and Purugganan 2018 ; Choi and Lee 2020 ]). Given the abundance of epigenetic variation in nature, opportunities may exist for genome-epigenome interactions involving transposable elements, leading to meaningful variation of phenotypes for adaptation.
I thank the members of the Yi lab for their comments and discussions. I apologize for authors whose articles that I did not have the space to cite. My research is partially supported by NSF (EF-2021635) and NIH (HG011641, E01MH134809) grants.
There is no new data associated with this article.
Alabert C , Jasencakova Z , Groth A . Chromatin replication and histone dynamics. In: Masai H , Foiani M , editors. DNA Replication. Advances in experimental medicine and biology . Singapore : Springer ; 2017 . p. 311 – 333 .
Google Scholar
Google Preview
Al Adhami H , Bardet AF , Dumas M , Cleroux E , Guibert S , Fauque P , Acloque H , Weber M . A comparative methylome analysis reveals conservation and divergence of DNA methylation patterns and functions in vertebrates . BMC Biol . 2022 : 20 ( 1 ): 70 . https://doi.org/10.1186/s12915-022-01270-x .
Allis CD , Jenuwein T . The molecular hallmarks of epigenetic control . Nat Rev Genet . 2016 : 17 ( 8 ): 487 – 500 . https://doi.org/10.1038/nrg.2016.59 .
Ammar R , Torti D , Tsui K , Gebbia M , Durbic T , Bader GD , Giaever G , et al. Chromatin is an ancient innovation conserved between Archaea and Eukarya . eLife . 2012 : 1 : e00078 . https://doi.org/10.7554/eLife.00078 .
Amukamara AU , Washington JT , Sanchez Z , McKinney EC , Moore AJ , Schmitz RJ , Moore PJ. More than DNA methylation: does pleiotropy drive the complex pattern of evolution of Dnmt1? Front Ecol Evol . 2020 : 8 : 4 . https://doi.org/10.3389/fevo.2020.00004 .
Angeloni A , Fissette S , Kaya D , Hammond JM , Gamaarachchi H , Deveson IW , Klose RJ , Li W , Zhang X , Bogdanovic O . Extensive DNA methylome rearrangement during early lamprey embryogenesis . Nat Commun . 2024 : 15 ( 1 ): 1977 . https://doi.org/10.1038/s41467-024-46085-2 .
Anway MD , Cupp AS , Uzumcu M , Skinner MK . Epigenetic transgenerational actions of endocrine disruptors and male fertility . Science . 2005 : 308 ( 5727 ): 1466 – 1469 . https://doi.org/10.1126/science.1108190 .
Arsala D , Wu X , Yi SV , Lynch JA . Dnmt1a is essential for gene body methylation and the regulation of the zygotic genome in a wasp . PLoS Genet. 2022 : 18 ( 5 ): e1010181 . https://doi.org/10.1371/journal.pgen.1010181 .
Ashe A , Sapetschnig A , Weick E-M , Mitchell J , Marloes PB , Amy CC , Doebley A-L , Leonard DG , Nicolas JL , Le Pen J , et al. piRNAs can trigger a multigenerational epigenetic memory in the Germline of C. elegans . Cell . 2012 : 150 ( 1 ): 88 – 99 . https://doi.org/10.1016/j.cell.2012.06.018 .
Beck D , Sadler-Riggleman I , Skinner MK . Generational comparisons (F1 versus F3) of vinclozolin induced epigenetic transgenerational inheritance of sperm differential DNA methylation regions (epimutations) using MeDIP-Seq . Environ Epigenet . 2017 : 3 ( 3 ): dvx016 . https://doi.org/10.1093/eep/dvx016 .
Becker C , Hagmann J , Muller J , Koenig D , Stegle O , Borgwardt K , Weigel D . Spontaneous epigenetic variation in the Arabidopsis thaliana methylome . Nature . 2011 : 480 ( 7376 ): 245 – 249 . https://doi.org/10.1038/nature10555 .
Beltran T , Shahrezaei V , Katju V , Sarkies P . Epimutations driven by small RNAs arise frequently but most have limited duration in Caenorhabditis elegans . Nat Ecol Evol . 2020 : 4 ( 11 ): 1539 – 1548 . https://doi.org/10.1038/s41559-020-01293-z .
Bertozzi TM , Ferguson-Smith AC . Metastable epialleles and their contribution to epigenetic inheritance in mammals . Semin Cell Dev Biol . 2020 : 97 : 93 – 105 . https://doi.org/10.1016/j.semcdb.2019.08.002 .
Bewick AJ , Ji L , Niederhuth CE , Willing E-M , Hofmeister BT , Shi X , Wang L , Lu Z , Rohr NA , Hartwig B , et al. On the origin and evolutionary consequences of gene body DNA methylation . Proc Natl Acad Sci U S A. 2016 : 113 ( 32 ): 9111 . https://doi.org/10.1073/pnas.1604666113 .
Bewick AJ , Sanchez Z , Mckinney EC , Moore AJ , Moore PJ , Schmitz RJ . Dnmt1 is essential for egg production and embryo viability in the large milkweed bug, Oncopeltus fasciatus . Epigenetics Chromatin . 2019 : 12 ( 1 ): 6 . https://doi.org/10.1186/s13072-018-0246-5 .
Bewick AJ , Schmitz RJ . Gene body DNA methylation in plants . Curr Opin Plant Biol . 2017 : 36 : 103 – 110 . https://doi.org/10.1016/j.pbi.2016.12.007 .
Bewick AJ , Vogel KJ , Moore AJ , Schmitz RJ . Evolution of DNA Methylation across insects . Mol Biol Evol . 2017 : 34 ( 3 ): 654 – 665 . https://doi.org/10.1093/molbev/msw264 .
Blake LE , Roux J , Hernando-Herraez I , Banovich NE , Perez RG , Hsiao CJ , Eres I , Cuevas C , Marques-Bonet T , Gilad Y . A comparison of gene expression and DNA methylation patterns across tissues and species . Genome Res . 2020 : 30 ( 2 ): 250 – 262 . https://doi.org/10.1101/gr.254904.119 .
Bogan SN , Yi SV . Potential role of DNA methylation as a driver of plastic responses to the environment across cells, organisms, and populations . Genome Biol Evol. 2024 : 16 ( 2 ): evae022 . https://doi.org/10.1093/gbe/evae022 .
Bossdorf O , Arcuri D , Richards CL , Pigliucci M . Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana . Evol Ecol . 2010 : 24 ( 3 ): 541 – 553 . https://doi.org/10.1007/s10682-010-9372-7 .
Bossdorf O , Richards CL , Pigliucci M . Epigenetics for ecologists . Ecol Lett . 2008 : 11 ( 2 ): 106 – 115 . https://doi.org/10.1111/j.1461-0248.2007.01130.x .
Brickner JH . Inheritance of epigenetic transcriptional memory through read–write replication of a histone modification . Ann N Y Acad Sci . 2023 : 1526 ( 1 ): 50 – 58 . https://doi.org/10.1111/nyas.15033 .
Brink K , Thomas CL , Jones A , Chan TW , Mallon EB . Exploring the ageing methylome in the model insect, Nasonia vitripennis . BMC Genom . 2024 : 25 ( 1 ): 305 . https://doi.org/10.1186/s12864-024-10211-7 .
Caglayan E , Ayhan F , Liu Y , Vollmer RM , Oh E , Sherwood CC , Preuss TM , Yi SV , Konopka G . Molecular features driving cellular complexity of human brain evolution . Nature . 2023 : 620 ( 7972 ): 145 – 153 . https://doi.org/10.1038/s41586-023-06338-4 .
Carja O , MacIsaac JL , Mah SM , Henn BM , Kobor MS , Feldman MW , Fraser HB . Worldwide patterns of human epigenetic variation . Nat Ecol Evol . 2017 : 1 ( 10 ): 1577 – 1583 . https://doi.org/10.1038/s41559-017-0299-z .
Cavalli G , Misteli T . Functional implications of genome topology . Nat Struct Mol Biol . 2013 : 20 ( 3 ): 290 – 299 . https://doi.org/10.1038/nsmb.2474 .
Charlesworth D , Barton NH , Charlesworth B . The sources of adaptive variation . Proc R Soc B Biol Sci . 2017 : 284 ( 1855 ): 20162864 . https://doi.org/10.1098/rspb.2016.2864 .
Chen L , Ge B , Casale FP , Vasquez L , Kwan T , Garrido-Martín D , Watt S , Yan Y , Kundu K , Ecker S , et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells . Cell . 2016 : 167 ( 5 ): 1398 – 1414.e1324 . https://doi.org/10.1016/j.cell.2016.10.026 .
Chen S , Liu S , Shi S , Yin H , Tang Y , Zhang J , Li W , Liu G , Qu K , Ding X , et al. Cross-species comparative DNA methylation reveals novel insights into complex trait genetics among cattle, sheep, and goats . Mol Biol Evol . 2024 : 41 ( 2 ): msae003 . https://doi.org/10.1093/molbev/msae003 .
Choi JY , Lee YCG . Double-edged sword: the evolutionary consequences of epigenetic silencing of transposable elements . PLoS Genet . 2020 : 16 ( 7 ): e1008872 . https://doi.org/10.1371/journal.pgen.1008872 .
Choi JY , Purugganan MD . Evolutionary epigenomics of retrotransposon-mediated methylation spreading in rice . Mol Biol Evol . 2018 : 35 ( 2 ): 365 – 382 . https://doi.org/10.1093/molbev/msx284 .
Cooney CA , Dave AA , Wolff GL . Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring . J Nutr . 2002 : 132 ( 8 ): 2393S – 2400S . https://doi.org/10.1093/jn/132.8.2393S .
Cropley JE , Suter CM , Beckman KB , Martin DI . Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation . Proc Natl Acad Sci U S A . 2006 : 103 ( 46 ): 17308 – 17312 . https://doi.org/10.1073/pnas.0607090103 .
Cropley JE , Suter CM , Beckman KB , Martin DIK . Cpg methylation of a silent controlling element in the murine avy allele is incomplete and unresponsive to methyl donor supplementation . PLoS One . 2010 : 5 ( 2 ): e9055 . https://doi.org/10.1371/journal.pone.0009055 .
Cusanovich DA , Hill AJ , Aghamirzaie D , Daza RM , Pliner HA , Berletch JB , Filippova GN , Huang X , Christiansen L , DeWitt WS , et al. A single-cell atlas of in vivo mammalian chromatin accessibility . Cell . 2018 : 174 ( 5 ): 1309 – 1324.e1318 . https://doi.org/10.1016/j.cell.2018.06.052 .
Daxinger L , Whitelaw E . Understanding transgenerational epigenetic inheritance via the gametes in mammals . Nat Rev Genet . 2012 : 13 ( 3 ): 153 – 162 . https://doi.org/10.1038/nrg3188 .
de Mendoza A , Lister R , Bogdanovic O . Evolution of DNA methylome diversity in eukaryotes . J Mol Biol . 2020 : 432 ( 6 ): 1687 – 1705 . https://doi.org/10.1016/j.jmb.2019.11.003 .
Deans C , Maggert KA . What do you mean, “epigenetic”? Genetics . 2015 : 199 ( 4 ): 887 – 896 . https://doi.org/10.1534/genetics.114.173492 .
Do C , Shearer A , Suzuki M , Terry MB , Gelernter J , Greally JM , Tycko B . Genetic–epigenetic interactions in cis: a major focus in the post-GWAS era . Genome Biol . 2017 : 18 ( 1 ): 120 . https://doi.org/10.1186/s13059-017-1250-y .
Duhl DMJ , Vrieling H , Miller KA , Wolff GL , Barsh GS . Neomorphic agouti mutations in obese yellow mice . Nat Genet . 1994 : 8 ( 1 ): 59 – 65 . https://doi.org/10.1038/ng0994-59 .
Duncan EJ , Cunningham CB , Dearden PK . Phenotypic plasticity: what has DNA methylation got to do with it? Insects . 2022 : 13 ( 2 ): 110 . https://doi.org/10.3390/insects13020110 .
Duncan EJ , Gluckman PD , Dearden PK . Epigenetics, plasticity, and evolution: how do we link epigenetic change to phenotype? J Exp Zool B Mol Dev Evol . 2014 : 322 ( 4 ): 208 – 220 . https://doi.org/10.1002/jez.b.22571 .
Eichten SR , Briskine R , Song J , Li Q , Swanson-Wagner R , Hermanson PJ , Waters AJ , Starr E , West PT , Tiffin P , et al. Epigenetic and genetic influences on DNA methylation variation in maize populations . Plant Cell . 2013 : 25 ( 8 ): 2783 – 2797 . https://doi.org/10.1105/tpc.113.114793 .
Erives AJ . Phylogenetic analysis of the core histone doublet and DNA topo II genes of marseilleviridae: evidence of proto-eukaryotic provenance . Epigenetics Chromatin . 2017 : 10 ( 1 ): 55 . https://doi.org/10.1186/s13072-017-0162-0 .
Feinberg AP . Phenotypic plasticity and the epigenetics of human disease . Nature . 2007 : 447 ( 7143 ): 433 – 440 . https://doi.org/10.1038/nature05919 .
Fitz-James MH , Cavalli G . Molecular mechanisms of transgenerational epigenetic inheritance . Nat Rev Genet . 2022 : 23 ( 6 ): 325 – 341 . https://doi.org/10.1038/s41576-021-00438-5 .
Fu F-F , Dawe RK , Gent JI . Loss of RNA-directed DNA methylation in maize chromomethylase and DDM1-type nucleosome remodeler mutants . Plant Cell . 2018 : 30 ( 7 ): 1617 – 1627 . https://doi.org/10.1105/tpc.18.00053 .
Ganguly DR , Crisp PA , Eichten SR , Pogson BJ . The Arabidopsis DNA methylome is stable under transgenerational drought stress . Plant Physiol . 2017 : 175 ( 4 ): 1893 – 1912 . https://doi.org/10.1104/pp.17.00744 .
Gao F , Liu XS , Wu X-P , Wang X-L , Gong D , Lu H , Song Y , Wang J , Du J , Liu S , et al. Differential methylation in discrete developmental stages of the parasitic nematode Trichinella spiralis . Genome Biol . 2012 : 13 ( 10 ): R100 . https://doi.org/10.1186/gb-2012-13-10-r100 .
Gao X , Thomsen H , Zhang Y , Breitling LP , Brenner H . The impact of methylation quantitative trait loci (mQTLs) on active smoking-related DNA methylation changes . Clin Epigenetics . 2017 : 9 ( 1 ): 87 . https://doi.org/10.1186/s13148-017-0387-6 .
García-Pérez R , Esteller-Cucala P , Mas G , Lobón I , Di Carlo V , Riera M , Kuhlwilm M , Navarro A , Blancher A , Di Croce L , et al. Epigenomic profiling of primate lymphoblastoid cell lines reveals the evolutionary patterns of epigenetic activities in gene regulatory architectures . Nat Commun . 2021 : 12 ( 1 ): 3116 . https://doi.org/10.1038/s41467-021-23397-1 .
Glastad KM , Hunt BG , Yi SV , Goodisman MAD . Epigenetic inheritance and genome regulation: is DNA methylation linked to ploidy in haplodiploid insects? Proc Biol Sci . 2014 : 281 ( 1785 ): 20140411 . https://doi.org/10.1098/rspb.2014.0411 .
Goll MG , Bestor TH . Eukaryotic cytosine methyltransferases . Annu Rev Biochem . 2005 : 74 ( 1 ): 481 – 514 . https://doi.org/10.1146/annurev.biochem.74.010904.153721 .
Grau-Bové X , Navarrete C , Chiva C , Pribasnig T , Antó M , Torruella G , Galindo LJ , Lang BF , Moreira D , López-Garcia P , et al. A phylogenetic and proteomic reconstruction of eukaryotic chromatin evolution . Nat Ecol Evol . 2022 : 6 ( 7 ): 1007 – 1023 . https://doi.org/10.1038/s41559-022-01771-6 .
Grbic M , Van Leeuwen T , Clark RM , Rombauts S , Rouze P , Grbic V , Osborne EJ , Dermauw W , Thi Ngoc PC , Ortego F , et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations . Nature . 2011 : 479 ( 7374 ): 487 – 492 . https://doi.org/10.1038/nature10640 .
Hämälä T , Ning W , Kuittinen H , Aryamanesh N , Savolainen O . Environmental response in gene expression and DNA methylation reveals factors influencing the adaptive potential of Arabidopsis lyrata . eLife . 2022 : 11 : e83115 . https://doi.org/10.7554/eLife.83115 .
Hanson MA , Skinner MK . Developmental origins of epigenetic transgenerational inheritance . Environ Epigenet . 2016 : 2 ( 1 ): dvw002 . https://doi.org/10.1093/eep/dvw002 .
He L , Huang H , Bradai M , Zhao C , You Y , Ma J , Zhao L , Lozano-Durán R , Zhu J-K . DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development . Nat Commun . 2022 : 13 ( 1 ): 1335 . https://doi.org/10.1038/s41467-022-28940-2 .
Heard E , Martienssen RA . Transgenerational epigenetic inheritance: myths and mechanisms . Cell . 2014 : 157 ( 1 ): 95 – 109 . https://doi.org/10.1016/j.cell.2014.02.045 .
Hellman A , Chess A . Gene body-specific methylation on the active X chromosome . Science . 2007 : 315 ( 5815 ): 1141 – 1143 . https://doi.org/10.1126/science.1136352 .
Henderson IR , Jacobsen SE . Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading . Genes Dev . 2008 : 22 ( 12 ): 1597 – 1606 . https://doi.org/10.1101/gad.1667808 .
Hollister JD , Gaut BG . Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression . Genome Res . 2009 : 19 ( 8 ): 1419 – 1428 . https://doi.org/10.1101/gr.091678.109 .
Hu L , Li N , Xu C , Zhong S , Lin X , Yang J , Zhou T , Yuliang A , Wu Y , Chen Y-R , et al. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality . Proc Natl Acad Sci U S A . 2014 : 111 ( 29 ): 10642 – 10647 . https://doi.org/10.1073/pnas.1410761111 .
Hu Y , Yuan S , Du X , Liu J , Zhou W , Wei F . Comparative analysis reveals epigenomic evolution related to species traits and genomic imprinting in mammals . Innovation (Camb) . 2023 : 4 ( 3 ): 100434 . https://doi.org/10.1016/j.xinn.2023.100434 .
Huh I , Zeng J , Park T , Yi S . DNA methylation and transcriptional noise . Epigenetics Chromatin . 2013 : 6 ( 1 ): 9 . https://doi.org/10.1186/1756-8935-6-9 .
Hunt BG , Glastad KM , Yi SV , Goodisman MAD . The function of intragenic DNA methylation: insights from insect epigenomes . Integr Comp Biol . 2013 : 53 ( 2 ): 319 – 328 . https://doi.org/10.1093/icb/ict003 .
Inawaka K , Kawabe M , Takahashi S , Doi Y , Tomigahara Y , Tarui H , Abe J , Kawamura S , Shirai T . Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations . Toxicol Appl Pharmacol . 2009 : 237 ( 2 ): 178 – 187 . https://doi.org/10.1016/j.taap.2009.03.004 .
Ivasyk I , Olivos-Cisneros L , Valdés-Rodríguez S , Droual M , Jang H , Schmitz RJ , Kronauer DJC . DNMT1 mutant ants develop normally but have disrupted oogenesis . Nat Commun . 2023 : 14 ( 1 ): 2201 . https://doi.org/10.1038/s41467-023-37945-4 .
Jablonka E , Raz G . Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution . Q Rev Biol . 2009 : 84 ( 2 ): 131 – 176 . https://doi.org/10.1086/598822 .
Jeong H , Mendizabal I , Berto S , Chatterjee P , Layman T , Usui N , Toriumi K , Douglas C , Singh D , Huh I , et al. Evolution of DNA methylation in the human brain . Nat Commun . 2021 : 12 ( 1 ): 2021 . https://doi.org/10.1038/s41467-021-21917-7 .
Jeong H , Wu X , Smith B , Yi SV . Genomic landscape of methylation islands in hymenopteran insects . Genome Biol Evol . 2018 : 10 ( 10 ): 2766 – 2776 . https://doi.org/10.1093/gbe/evy203 .
Jiang C , Mithani A , Belfield EJ , Mott R , Hurst LD , Harberd NP . Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations . Genome Res . 2014 : 24 ( 11 ): 1821 – 1829 . https://doi.org/10.1101/gr.177659.114 .
Johannes F , Porcher E , Teixeira FK , Saliba-Colombani V , Simon M , Agier N , As B , Albuisson J , Heredia F , Audigier P , et al. Assessing the impact of transgenerational epigenetic variation on complex traits . PLoS Genet . 2009 : 5 ( 6 ): e1000530 . https://doi.org/10.1371/journal.pgen.1000530 .
Jones PA . Functions of DNA methylation: islands, start sites, gene bodies and beyond . Nat Rev Genet . 2012 : 201 ( 7 ): 484 – 492 . https://doi.org/10.1038/nrg3230 .
Kankel MW , Ramsey DE , Stokes TL , Flowers SK , Haag JR , Jeddeloh JA , Riddle NC , Verbsky ML , Richards EJ . Arabidopsis MET1 cytosine methyltransferase mutants . Genetics . 2003 : 163 ( 3 ): 1109 – 1122 . https://doi.org/10.1093/genetics/163.3.1109 .
Keller TE , Han P , Yi SV . Evolutionary transition of promoter and gene body DNA methylation across invertebrate-vertebrate boundary . Mol Biol Evol. 2015 : 33 ( 4 ): 1019 – 1028 . https://doi.org/10.1093/molbev/msv345 .
Klughammer J , Romanovskaia D , Nemc A , Posautz A , Seid CA , Schuster LC , Keinath MC , Ramos JSL , Kosack L , Evankow A , et al. Comparative analysis of genome-scale, base-resolution DNA methylation profiles across 580 animal species . Nat Commun . 2023 : 14 ( 1 ): 232 . https://doi.org/10.1038/s41467-022-34828-y .
Krebs AR , Dessus-Babus S , Burger L , Schübeler D . High-throughput engineering of a mammalian genome reveals building principles of methylation states at CG rich regions . eLife . 2014 : 3 : e04094 . https://doi.org/10.7554/eLife.04094 .
Kucharski R , Maleszka J , Foret S , Maleszka R . Nutritional control of reproductive status in honeybees via DNA methylation . Science . 2008 : 319 ( 5871 ): 1827 – 1830 . https://doi.org/10.1126/science.1153069 .
Kundaje A , Meuleman W , Ernst J , Bilenky M , Yen A , Heravi-Moussavi A , Kheradpour P , Zhang Z , Wang J , Ziller MJ , et al. Integrative analysis of 111 reference human epigenomes . Nature . 2015 : 518 ( 7539 ): 317 – 330 . https://doi.org/10.1038/nature14248 .
Latzel V , Allan E , Silveira AB , Colot V , Fischer M , Bossdorf O . Epigenetic diversity increases the productivity and stability of plant populations . Nat Commun . 2013 : 4 ( 1 ): 2875 . https://doi.org/10.1038/ncomms3875 .
Lee KS , Chatterjee P , Choi E-Y , Sung MK , Oh J , Won H , Park S-M , Kim Y-J , Yi SV , Choi JK . Selection on the regulation of sympathetic nervous activity in humans and chimpanzees . PLoS Genet . 2018 : 14 ( 4 ): e1007311 . https://doi.org/10.1371/journal.pgen.1007311 .
Li E , Bestor TH , Jaenisch R . Targeted mutation of the DNA methyltransferase gene results in embryonic lethality . Cell . 1992 : 69 ( 6 ): 915 – 926 . https://doi.org/10.1016/0092-8674(92)90611-F .
Liang W , Li J , Sun L , Liu Y , Lan Z , Qian W . Deciphering the synergistic and redundant roles of CG and non-CG DNA methylation in plant development and transposable element silencing . New Phytol . 2022 : 233 ( 2 ): 722 – 737 . https://doi.org/10.1111/nph.17804 .
Liao J , Karnik R , Gu H , Ziller MJ , Clement K , Tsankov AM , Akopian V , Gifford CA , Donaghey J , Galonska C , et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells . Nat Genet . 2015 : 47 ( 5 ): 469 – 478 . https://doi.org/10.1038/ng.3258 .
Lienert F , Wirbelauer C , Som I , Dean A , Mohn F , Schubeler D . Identification of genetic elements that autonomously determine DNA methylation states . Nat Genet . 2011 : 43 ( 11 ): 1091 – 1097 . https://doi.org/10.1038/ng.946 .
Long HK , Prescott SL , Wysocka J . Ever-changing landscapes: transcriptional enhancers in development and evolution . Cell . 2016 : 167 ( 5 ): 1170 – 1187 . https://doi.org/10.1016/j.cell.2016.09.018 .
Loughland I , Little A , Seebacher F . DNA methyltransferase 3a mediates developmental thermal plasticity . BMC Biol . 2021 : 19 ( 1 ): 11 . https://doi.org/10.1186/s12915-020-00942-w .
Lu AT , Fei Z , Haghani A , Robeck TR , Zoller JA , Li CZ , Lowe R , Yan Q , Zhang J , Vu H , et al. Universal DNA methylation age across mammalian tissues . Nat Aging . 2023 : 3 ( 9 ): 1144 – 1166 . https://doi.org/10.1038/s43587-023-00462-6 .
Lyko F . The DNA methyltransferase family: a versatile toolkit for epigenetic regulation . Nat Rev Genet . 2018 : 19 ( 2 ): 81 – 92 . https://doi.org/10.1038/nrg.2017.80 .
Makova KD , Hardison RC . The effects of chromatin organization on variation of mutation rates in the genome . Nat Rev Genet . 2015 : 16 ( 4 ): 213 – 223 . https://doi.org/10.1038/nrg3890 .
Mathieu O , Reinders J , Caikovsk M , Smathajitt C , Paszkowski J . Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation . Cell . 2007 : 5 ( 5 ): 851 – 862 . https://doi.org/10.1016/j.cell.2007.07.007 .
Mendizabal I , Berto S , Usui N , Toriumi K , Chatterjee P , Douglas C , Huh I , Jeong H , Layman T , Tamminga CA , et al. Cell type-specific epigenetic links to schizophrenia risk in the brain . Genome Biol . 2019 : 20 ( 1 ): 135 . https://doi.org/10.1186/s13059-019-1747-7 .
Mendizabal I , Shi L , Keller TE , Konopka G , Preuss TM , Hsieh T-F , Hu E , Zhang Z , Su B , Yi SV . Comparative methylome analyses identify epigenetic regulatory loci of human brain evolution . Mol Biol Evol . 2016 : 33 ( 11 ): 2947 – 2959 . https://doi.org/10.1093/molbev/msw176 .
Mendizabal I , Zeng J , Keller TE , Yi SV . Body-hypomethylated human genes harbor extensive intragenic transcriptional activity and are prone to cancer-associated dysregulation . Nucl Acids Res . 2017 : 45 ( 8 ): 4390 – 4400 . https://doi.org/10.1093/nar/gkx020 .
Molaro A , Hodges E , Fang F , Song Q , McCombie WR , Hannon GJ , Smith AD . Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates . Cell . 2011 : 146 ( 6 ): 1029 – 1041 . https://doi.org/10.1016/j.cell.2011.08.016 .
Morgan HD , Sutherland HGE , Martin DIK , Whitelaw E . Epigenetic inheritance at the agouti locus in the mouse . Nat Genet . 1999 : 23 ( 3 ): 314 – 318 . https://doi.org/10.1038/15490 .
Morgan R , Loh E , Singh D , Mendizabal I , Yi SV. 2024 . DNA methylation differences between the female and male X chromosomes in human brain . bioRxiv 589778 . https://doi.org/10.1101/2024.04.16.589778 , 17 Aril 2024 , preprint: not peer reviewed.
Muyle A , Ross-Ibarra J , Seymour DK , Gaut BS . Gene body methylation is under selection in Arabidopsis thaliana . Genetics . 2021 : 218 ( 2 ): iyab061 . https://doi.org/10.1093/genetics/iyab061 .
Noshay JM , Springer NM . Stories that can’t be told by SNPs; DNA methylation variation in plant populations . Curr Opin Plant Biol . 2021 : 61 : 101989 . https://doi.org/10.1016/j.pbi.2020.101989 .
Okano M , Bell DW , Haber DA , Li E . DNA methyltransferases Dnmt3a and Dnmt3b are essential for De Novo methylation and mammalian development . Cell . 1999 : 99 ( 3 ): 247 – 257 . https://doi.org/10.1016/S0092-8674(00)81656-6 .
Pal S , Tyler JK . Epigenetics and aging . Sci Adv . 2016 : 2 ( 7 ): e1600584 . https://doi.org/10.1126/sciadv.1600584 .
Petronis A . Epigenetics as a unifying principle in the aetiology of complex traits and diseases . Nature . 2010 : 465 ( 7299 ): 721 – 727 . https://doi.org/10.1038/nature09230 .
Probst AV , Dunleavy E , Almouzni G . Epigenetic inheritance during the cell cycle . Nat Rev Mol Cell Biol . 2009 : 10 ( 3 ): 192 – 206 . https://doi.org/10.1038/nrm2640 .
Rechavi O , Houri-Ze’evi L , Anava S , Goh Wee Siong S , Sze YK , Gregory JH , Hobert O . Starvation-Induced transgenerational inheritance of small RNAs in C. elegans . Cell . 2014 : 158 ( 2 ): 277 – 287 . https://doi.org/10.1016/j.cell.2014.06.020 .
Rechavi O , Lev I . Principles of transgenerational small RNA inheritance in Caenorhabditis elegans . Curr Biol . 2017 : 27 ( 14 ): R720 – R730 . https://doi.org/10.1016/j.cub.2017.05.043 .
Richards CL , Alonso C , Becker C , Bossdorf O , Bucher E , Colomé-Tatché M , Durka W , Engelhardt J , Gaspar B , Gogol-Döring A , et al. Ecological plant epigenetics: evidence from model and non-model species, and the way forward . Ecol Lett . 2017 : 20 ( 12 ): 1576 – 1590 . https://doi.org/10.1111/ele.12858 .
Roberts SB , Gavery MR . Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front Physiol . 2012 : 2 : 116 . https://doi.org/10.3389/fphys.2011.00116 .
Sadler KC . Epigenetics across the evolutionary tree: new paradigms from non-model animals . BioEssays . 2023 : 45 ( 1 ): e2200036 . https://doi.org/10.1002/bies.202200036 .
Sandman K , Reeve JN . Archaeal histones and the origin of the histone fold . Curr Opin Microbiol . 2006 : 9 ( 5 ): 520 – 525 . https://doi.org/10.1016/j.mib.2006.08.003 .
Sarkies P . Encyclopaedia of eukaryotic DNA methylation: from patterns to mechanisms and functions . Biochem Soc Transact . 2022 : 50 : 1179 – 1190 . https://doi.org/10.1042/BST20210725 .
Schmitz RJ , Lewis ZA , Goll MG . DNA methylation: shared and divergent features across eukaryotes . Trends Genet . 2019 : 35 ( 11 ): 818 – 827 . https://doi.org/10.1016/j.tig.2019.07.007 .
Schmitz RJ , Schultz MD , Urich MA , Nery JR , Pelizzola M , Libiger O , Alix A , McCosh RB , Chen H , Schork NJ , et al. Patterns of population epigenomic diversity . Nature . 2013 : 495 ( 7440 ): 193 – 198 . https://doi.org/10.1038/nature11968 .
Schneider S , Kaufmann W , Buesen R , van Ravenzwaay B . Vinclozolin—the lack of a transgenerational effect after oral maternal exposure during organogenesis . Reprod Toxicol . 2008 : 25 ( 3 ): 352 – 360 . https://doi.org/10.1016/j.reprotox.2008.04.001 .
Schneider S , Marxfeld H , Gröters S , Buesen R , van Ravenzwaay B . Vinclozolin—no transgenerational inheritance of anti-androgenic effects after maternal exposure during organogenesis via the intraperitoneal route . Reprod Toxicol . 2013 : 37 : 6 – 14 . https://doi.org/10.1016/j.reprotox.2012.12.003 .
Schübeler D . Function and information content of DNA methylation . Nature . 2015 : 517 ( 7534 ): 321 – 326 . https://doi.org/10.1038/nature14192 .
Sepers B , Chen RS , Memelink M , Verhoeven KJF , van Oers K . Variation in DNA methylation in avian nestlings is largely determined by genetic effects . Mol Biol Evol . 2023 : 40 ( 4 ): msad086 . https://doi.org/10.1093/molbev/msad086 .
Seymour DK , Becker C . The causes and consequences of DNA methylome variation in plants . Curr Opin Plant Biol . 2017 : 36 : 56 – 63 . https://doi.org/10.1016/j.pbi.2017.01.005 .
Seymour DK , Gaut BS . Phylogenetic shifts in gene body methylation correlate with gene expression and reflect trait conservation . Mol Biol Evol . 2020 : 37 ( 1 ): 31 – 43 . https://doi.org/10.1093/molbev/msz195 .
Shi L , Lin Q , Su B . Human-Specific hypomethylation of CENPJ, a key brain size regulator . Mol Biol Evol . 2014 : 31 ( 3 ): 594 – 604 . https://doi.org/10.1093/molbev/mst231 .
Singh D , Sun D , King AG , Alquezar-Planas DE , Johnson RN , Alvarez-Ponce D , Yi SV . Koala methylomes reveal divergent and conserved DNA methylation signatures of X chromosome regulation . Proc R Soc B Biol Sci . 2021 : 288 ( 1945 ): 20202244 . https://doi.org/10.1098/rspb.2020.2244 .
Springer NM , Schmitz RJ . Exploiting induced and natural epigenetic variation for crop improvement . Nat Rev Genet . 2017 : 18 ( 9 ): 563 – 575 . https://doi.org/10.1038/nrg.2017.45 .
Stroud H , Do T , Du J , Zhong X , Feng S , Johnson L , Patel DJ , Jacobsen SE . Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis . Nat Struct Mol Biol . 2014 : 21 ( 1 ): 64 – 72 . https://doi.org/10.1038/nsmb.2735 .
Suzuki MM , Bird A . DNA methylation landscapes: provocative insights from epigenomics . Nat Rev Genet . 2008 : 9 ( 6 ): 465 – 476 . https://doi.org/10.1038/nrg2341 .
Takahashi Y , Valencia MM , Yu Y , Ouchi Y , Takahashi K , Shokhirev MN , Lande K , Williams AE , Fresia C , Kurita M , et al. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice . Cell . 2023 : 186 ( 4 ): 715 – 731 . https://doi.org/10.1016/j.cell.2022.12.047 .
Takuno S , Gaut BG . Gene body methylation is conserved between plant orthologs and is of evolutionary consequence . Proc Natl Acad Sci U S A . 2013 : 110 ( 5 ): 1797 – 1802 . https://doi.org/10.1073/pnas.1215380110 .
Talbert PB , Meers MP , Henikoff S . Old cogs, new tricks: the evolution of gene expression in a chromatin context . Nat Rev Genet . 2019 : 20 ( 5 ): 283 – 297 . https://doi.org/10.1038/s41576-019-0105-7 .
Trowbridge JJ , Snow JW , Kim J , Orkin SH . DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells . Cell Stem Cell . 2009 : 5 ( 4 ): 442 – 449 . https://doi.org/10.1016/j.stem.2009.08.016 .
Tweedie S , Charlton J , Clark V , Bird A . Methylation of genomes and genes at the invertebrate-vertebrate boundary . Mol Cell Biol . 1997 : 17 ( 3 ): 1469 – 1475 . https://doi.org/10.1128/MCB.17.3.1469 .
van der Graaf A , Wardenaar R , Neumann DA , Taudt A , Shaw RG , Jansen RC , Schmitz RJ , Colomé-Tatché M , Johannes F . Rate, spectrum, and evolutionary dynamics of spontaneous epimutations . Proc Natl Acad Sci U S A . 2015 : 112 ( 21 ): 6676 – 6681 . https://doi.org/10.1073/pnas.1424254112 .
Van Dooren TJM , Silveira AB , Gilbault E , Jiménez-Gómez JM , Martin A , Bach L , Tisné S , Quadrana L , Loudet O , Colot V . Mild drought in the vegetative stage induces phenotypic, gene expression, and DNA methylation plasticity in Arabidopsis but no transgenerational effects . J Exp Bot . 2020 : 71 ( 12 ): 3588 – 3602 . https://doi.org/10.1093/jxb/eraa132 .
Venney CJ , Anastasiadi D , Wellenreuther M , Bernatchez L . The evolutionary complexities of DNA methylation in animals: from plasticity to genetic evolution . Genome Biol Evol . 2023 : 15 ( 12 ): evad216 . https://doi.org/10.1093/gbe/evad216 .
Vidalis A , Živković D , Wardenaar R , Roquis D , Tellier A , Johannes F . Methylome evolution in plants . Genome Biol . 2016 : 17 ( 1 ): 264 . https://doi.org/10.1186/s13059-016-1127-5 .
Villar D , Berthelot C , Aldridge S , Tim FR , Lukk M , Pignatelli M , Thomas JP , Deaville R , Erichsen Jonathan T , Anna JJ , et al. Enhancer evolution across 20 mammalian Species . Cell . 2015 : 160 ( 3 ): 554 – 566 . https://doi.org/10.1016/j.cell.2015.01.006 .
Villicaña S , Bell JT . Genetic impacts on DNA methylation: research findings and future perspectives . Genome Biol . 2021 : 22 ( 1 ): 127 . https://doi.org/10.1186/s13059-021-02347-6 .
Waddington CH . The strategy of the genes . London : Routledge ; 1957 .
Wang Y, Jorda M, Jones PL, Maleszka R, Ling X, Robertson HM, Mizzen CA, Peinado MA, Robinson GE. Functional CpG methylation system in a social insect. Sci. 2006: 314 :645–647.
Wang X, Werren JH, Clark AG . Allele-Specific Transcriptome and Methylome Analysis Reveals Stable Inheritance and Cis-Regulation of DNA Methylation in Nasonia . PLOS Biol . 2016 : 14 ( 7 ): e1002500 . https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002500 .
Washington JT , Cavender KR , Amukamara AU , McKinney EC , Schmitz RJ , Moore PJ . The essential role of dnmt1 in Gametogenesis in the large milkweed bug Oncopeltus fasciatus . eLife . 2021 : 10 : e62202 . https://doi.org/10.7554/eLife.62202 .
Waterland RA , Jirtle RL . Transposable elements: targets for early nutritional effects on epigenetic gene regulation . Mol Cell Biol . 2003 : 23 ( 15 ): 5293 – 5300 . https://doi.org/10.1128/MCB.23.15.5293-5300.2003 .
Weber M , Davies JJ , Wittig D , Oakeley EJ , Haase M , Lam WL , Schübeler D . Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells . Nat Genet . 2005 : 37 ( 8 ): 853 – 862 . https://doi.org/10.1038/ng1598 .
Werren JH , Richards S , Desjardins CA , Niehuis O , Gadau J , Colbourne JK ; Nasonia Genome Working Group ; Werren JH , Richards S , Desjardins CA . Functional and evolutionary insights from the genomes of three parasitoid Nasonia Species . Science . 2010 : 327 ( 5963 ): 343 – 348 . https://doi.org/10.1126/science.1178028 .
Wolff GL . Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse . Genetics . 1978 : 88 ( 3 ): 529 – 539 . https://doi.org/10.1093/genetics/88.3.529 .
Wolff GL , Kodell RL , Moore SR , Cooney CA . Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice . FASEB J . 1998 : 12 ( 11 ): 949 – 957 . https://doi.org/10.1096/fasebj.12.11.949 .
Wu X , Bhatia N , Grozinger CM , Yi SV . Comparative studies of genomic and epigenetic factors influencing transcriptional variation in two insect species . G3 (Bethesda) . 2022 : 12 ( 11 ): jkac230 . https://doi.org/10.1093/g3journal/jkac230 .
Wu X , Galbraith DA , Chatterjee P , Jeong H , Grozinger CM , Yi SV . Lineage and parent-of-origin effects in DNA methylation of honey bees ( Apis mellifera ) revealed by reciprocal crosses and whole-genome bisulfite sequencing . Genome Biol Evol . 2000 : 12 ( 8 ): 1482 – 1492 . https://doi.org/10.1093/gbe/evaa133 .
Yi SV . Birds do it, bees do it, worms and ciliates do it too: DNA methylation from unexpected corners of the tree of life . Genome Biol . 2012 : 13 ( 10 ): 174 . https://doi.org/10.1186/gb-2012-13-10-174 .
Yi SV . Insights into epigenome evolution from animal and plant methylomes . Genome Biol Evol . 2017 : 9 ( 11 ): 3189 – 3201 . https://doi.org/10.1093/gbe/evx203 .
Yi SV , Goodisman MAD . The impact of epigenetic information on genome evolution . Philos Trans R Soc B Biol Sci . 2021 : 376 ( 1826 ): 20200113 . https://doi.org/10.1098/rstb.2020.0114 .
Zeng J , Konopka G , Hunt BG , Preuss TM , Geschwind D , Yi SV . Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution . Am J Human Genet . 2012 : 91 ( 3 ): 455 – 465 . https://doi.org/10.1016/j.ajhg.2012.07.024 .
Zhang L , Lu Q , Chang C . Epigenetics in health and disease. In: Chang C , Lu Q , editors. Epigenetics in allergy and autoimmunity. Advances in experimental medicine and biology . 1253 . Singapore : Springer ; 2020 . p. 3 – 55 .
Zhang Y-Y , Fischer M , Colot V , Bossdorf O . Epigenetic variation creates potential for evolution of plant phenotypic plasticity . New Phytol . 2013 : 197 ( 1 ): 314 – 322 . https://doi.org/10.1111/nph.12010 .
Zhong X . Comparative epigenomics: a powerful tool to understand the evolution of DNA methylation . New Phytol . 2016 : 210 ( 1 ): 76 – 80 . https://doi.org/10.1111/nph.13540 .
Zilberman D . An evolutionary case for functional gene body methylation in plants and animals . Genome Biol . 2017 : 18 ( 1 ): 87 . https://doi.org/10.1186/s13059-017-1230-2 .
Email alerts
Citing articles via.
- Author Guidelines
Affiliations
- Online ISSN 1537-1719
- Copyright © 2024 Society for Molecular Biology and Evolution
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Biological Research
Special series on Microbial Interactions
The nine articles of this special issue of Biological Research address biochemical and genetic determinants of microbial response and tolerance to stressors in different biological models and environmental contexts. Individual articles provide a broad exploration of our current knowledge of response to stressors, with a special emphasis on metal metabolism and toxic compounds.

Special series on Antarctic Research
This special issue on Antarctic research in Biological Research comprises of recent studies, related to the discovery of several new enzymes and biotechnological applications that allow to expand the knowledge of Antarctic organisms and their potential applications.
- Most accessed
Fruit sugar hub: gene regulatory network associated with soluble solids content (SSC) in Prunus persica
Authors: Gerardo Núñez-Lillo, Victoria Lillo-Carmona, Alonso G. Pérez-Donoso, Romina Pedreschi, Reinaldo Campos-Vargas and Claudio Meneses
Antimicrobial activity of compounds identified by artificial intelligence discovery engine targeting enzymes involved in Neisseria gonorrhoeae peptidoglycan metabolism
Authors: Ravi Kant, Hannah Tilford, Camila S. Freitas, Dayana A. Santos Ferreira, James Ng, Gwennan Rucinski, Joshua Watkins, Ryan Pemberton, Tigran M. Abramyan, Stephanie C. Contreras, Alejandra Vera and Myron Christodoulides
Hormonal influence: unraveling the impact of sex hormones on vascular smooth muscle cells
Authors: Keran Jia, Xin Luo, Jingyan Yi and Chunxiang Zhang
Unraveling the impact of hyperleptinemia on female reproduction: insights from transgenic pig model
Authors: Muhammad Ameen Jamal, Yixiao Cheng, Deling Jiao, Wen Cheng, Di Zou, Xia Wang, Taiyun Wei, Jianxiong Guo, Kaixiang Xu, Heng Zhao, Shaoxia Pu, Chang Yang, Yubo Qing, Baoyu Jia, Honghui Li, Rusong Zhao…
Deciphering genetic and nongenetic factors underlying tumour dormancy: insights from multiomics analysis of two syngeneic MRD models of melanoma and leukemia
Authors: Marie-Océane Laguillaumie, Sofia Titah, Aurélie Guillemette, Bernadette Neve, Frederic Leprêtre, Pascaline Ségard, Faruk Azam Shaik, Dominique Collard, Jean-Claude Gerbedoen, Léa Fléchon, Lama Hasan Bou Issa, Audrey Vincent, Martin Figeac, Shéhérazade Sebda, Céline Villenet, Jérôme Kluza…
Most recent articles RSS
View all articles
Awareness and current knowledge of breast cancer
Authors: Muhammad Akram, Mehwish Iqbal, Muhammad Daniyal and Asmat Ullah Khan
Stress and defense responses in plant secondary metabolites production
Authors: Tasiu Isah
Fate of nitrogen in agriculture and environment: agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency
Authors: Muhammad Anas, Fen Liao, Krishan K. Verma, Muhammad Aqeel Sarwar, Aamir Mahmood, Zhong-Liang Chen, Qiang Li, Xu-Peng Zeng, Yang Liu and Yang-Rui Li
Coping with drought: stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness
Authors: Aziz Khan, Xudong Pan, Ullah Najeeb, Daniel Kean Yuen Tan, Shah Fahad, Rizwan Zahoor and Honghai Luo
Biotechnological applications of archaeal enzymes from extreme environments
Authors: Ma. Ángeles Cabrera and Jenny M. Blamey
Most accessed articles RSS
Aims and scope
Archival content.
Biological Research , formerly Archives of Experimental Medicine and Biology , was founded in 1964 and transferred to BioMed Central in 2014. An electronic archive of articles published between 1999 and 2013 can be found in the SciELO database.

Open access funding information
To find more information on research funders and institutions worldwide that fund open access article-processing charges (APCs) visit this page .
Latest Tweets
Your browser needs to have JavaScript enabled to view this timeline

Editor’s profile
Manuel J Santos, Editor-in-Chief
Dr Santos is an Associate Professor in the Faculty of Biological Sciences and Medicine at the Pontificia Catholic University of Chile.
Dr Santos received his MD from the University of Chile and his PhD in Cell and Molecular Biology from the Pontificia Catholic University of Chile. He majored in Medical Genetics at The John Hopkins University (USA) and The René Descartes University of Paris (France), and held a post doctorate position in Cell Biology and Genetics at the Rockefeller University (USA).
His research has focused on the biogenesis of cellular organelles, particularly peroxisomes. A pioneer in this field, his research lead him to discover a new type of human genetic disease, the peroxisomal biogenesis disorders, which include Zellweger Syndrome. More recently his research has centered on studying the role of peroxisomes in Alzheimer’s disease, and he also works in the field of bioethics.
Over the span of his career, Dr Santos has published more than 70 peer reviewed papers and been the President of the Society of Biology of Chile, the Genetics Society of Chile and the Bioethical Society of Chile.
About the Society
The Chilean Biology Society (Sociedad de Biología de Chile), previously the Biological Society of Santiago, was founded in late 1928 as a subsidiary of The Societé de Biologie of Paris, France. For several years the summaries of its communications were published in Comps Rendú of the Societé de Biologie du Paris. The Society is currently a member of the International Union of Biological Sciences (IUBS).
The Chilean Biology Society promotes theoretical and experimental studies and research leading to advancement in and dissemination of the biological sciences for the benefit of the community. To accomplish this, the Society organizes periodic scientific meetings in which scientists communicate, comment and discuss research carried out in Chilean or foreign research laboratories. In addition, relations and cooperation with similar domestic and foreign institutions are stimulated, and communication by all appropriate means of biological research carried out in Chile.
Members of the Society will receive a discount on Biological Research 's article-processing charge when they provide a discount code (which members can obtain by emailing the Society) during the submission process. The discounted article-processing charge for Society members is £1150 in 2023.
The Society also publishes Revista Chilena de Historia Natural ( Chilean Journal of Natural History, founded in 1897).

Annual Journal Metrics
Citation Impact 2023 Journal Impact Factor: 4.3 5-year Journal Impact Factor: 6.6 Source Normalized Impact per Paper (SNIP): 1.354 SCImago Journal Rank (SJR): 1.300 Speed 2023 Submission to first editorial decision (median days): 22 Submission to acceptance (median days): 155 Usage 2023 Downloads: 489,080 Altmetric mentions: 731
- More about our metrics
Official journal of

Chilean Biology Society ( Sociedad de Biología de Chile ).

- Editorial Board
- Manuscript editing services
- Instructions for Editors
- Sign up for article alerts and news from this journal
- Follow us on Twitter
- Follow us on Facebook
ISSN: 0717-6287
- Submission enquiries: Access here and click Contact Us
- General enquiries: [email protected]
Browsing: Biology
Biology is the scientific study of life and living organisms, encompassing various sub-disciplines such as microbiology, botany, zoology, and physiology. We’re dedicated to bringing you the latest research findings, innovative technologies, and thought-provoking discoveries from top scientists, research institutions, and universities around the world.
This section on biology news includes new research related to many related subjects such as biochemistry, genetics, cytology, and microbiology. Popular sub-topics include Biotechnology , DNA , Microbiology , Neurology , Evolutionary Biology , Genetics , Stem Cells , Neuroscience , Bioengineering , and Cell Biology .
Whether you are a professional biologist, an aspiring scientist, or simply someone with a passion for learning about the living world, our Biology News page offers a wealth of information and insights to keep you informed and inspired.
Mysterious Clones: Invasive Jellyfish From China Alarm British Columbia Researchers
Invasive peach blossom jellyfish, originating from China, have proliferated across British Columbia’s freshwater bodies, with…
Scientists Discover Peculiar New Species of Dragonfish in Antarctica
Newly identified Antarctic dragonfish, Akarotaxis gouldae, displays unique traits and faces threats from climate change…
Doubling Lifespan: Scientists Have Discovered a Key Cellular Mechanism That Could Control Longevity
UC Merced researchers have found that the protein OTUD6 can alter protein production in cells,…
Neural Surprises: Scientists Discover New Brain Mechanism
Researchers identify how brain circuits in mice amplify sensory prediction errors through a unique interaction…
Shocking Breakthrough: Newly Discovered Hybrid Brain Cells Fire Electrical Impulses
Scientists from Baylor College have identified a new cell type in the human brain that…
Scientists Discover Health-Boosting Probiotics in Brazillian Cheeses
In a new paper, researchers from Brazil discuss the benefits of three Lactobacillus bacteria strains…
“Astonishing” Finding: Genomic Dark Matter Solves Butterfly Evolutionary Mystery
New research uncovers a surprising genetic mechanism that impacts the development of butterfly wing colors…
The Surprising Origin of the Next Super Antibiotic: Your Gut
Researchers have discovered several new antibiotic candidates by mining gut microbiome data, with one matching…
Making the Neurodivergent Brain Visible: New Research Cracks the Autism Code
Researchers have developed a technique that accurately identifies genetic markers of autism in brain images,…
Scientists Discover Two Stunning New Fish Species in Pearl River
Two new species of hillstream suck-loach, Beaufortia granulopinna and Beaufortia viridis, discovered in China, exhibit…
New Research Suggests a Possible Prevention Path for Autism
Researchers found that altering neurotransmitters in early development could prevent autism spectrum disorders in mice,…
The Deadliest Predator: How Mosquitoes Use Infrared To Hunt Humans
UC Santa Barbara scientists found that Aedes aegypti mosquitoes use infrared radiation, in addition to…
Revolutionary DNA Study Maps the Genesis of Terrestrial Plant Life
Researchers have decoded the genomic sequence of Zygnema algae, revealing insights into the evolutionary transition…
When RNA Mistakes Turn Into Genetic Master Controls
New research from the University of Chicago reveals that alternative splicing plays a much bigger…
“Really Weird” – Deep-Sea Dark Oxygen Discovery Raises Questions About Alien Life
Researchers from Boston University have made a startling discovery: rocks are producing “dark oxygen” in…
Darwin’s Fear Was Unjustified: New Study Confirms Evolution Despite Fossil Gaps
Hiatuses in the rock record are not a significant issue. Fossils play a crucial role…
Unlocking the Secrets of Earth’s Most Resilient Creatures: New Fossils Reveal 500-Million-Year-Old History
Harvard scientists have discovered a new tardigrade species in amber, offering insights into their evolutionary…
How Simple Commands Turn Into Complex Behaviors – Optogenetics Reveals New Layers of Brain Function
A study on Drosophila reveals that descending neurons (DNs) responsible for behaviors form complex, behavior-specific…
Type above and press Enter to search. Press Esc to cancel.
An editorially independent publication supported by the Simons Foundation.
Get the latest news delivered to your inbox.
Type search term(s) and press enter
- Comment Comments
- Save Article Read Later Read Later
The Year in Biology
December 21, 2022

Myriam Wares for Quanta Magazine
Introduction
Our memories are the cornerstone of our identity. Their importance is a big part of what makes Alzheimer’s disease and other forms of dementia so cruel and poignant. It’s why we’ve hoped so desperately for science to deliver a cure for Alzheimer’s, and why it is so frustrating and tragic that useful treatments have been slow to emerge. Great excitement therefore surrounded the announcement in September that a new drug, lecanemab, slowed the progression of the disease in clinical trials. If it is approved by the Food and Drug Administration, lecanemab will become only the second Alzheimer’s treatment that counteracts amyloid-beta protein, which is widely supposed to be the cause of the disease.
Yet the effects of lecanemab are so marginal that researchers debate whether the drug will really make a practical difference for patients. The fact that lecanemab stands out as a bright spot speaks to how dismal much of the history of research on treatments for Alzheimer’s has been. Meanwhile, a deeper understanding of the biology at play is fueling interest in the leading alternative theories for what causes the disease.
Speculation about how memory works is at least as old as Plato, who in one of his Socratic dialogues wrote about “the gift of Memory, the mother of the Muses,” and compared its operation to a wax stamp in the soul. We can be grateful that science has vastly improved on our understanding of memory since Plato’s time — out with the wax stamps, in with “engrams” of changes in our neurons. In this past year alone, researchers have made exciting strides toward learning how and where in the brain different aspects of our memories reside. More surprisingly, they have even found biochemical mechanisms that distinguish good memories from bad ones.
Because we are creatures with brains, we often think about memory in purely neurological terms. Yet work published early in 2022 by researchers at the California Institute of Technology suggests that even individual cells in developing tissues may carry some records of their lineage’s history. These stem cells seem to rely on that stored information when they are faced with decisions about how to specialize in response to chemical cues. Advances in biology over this past year unveiled many other surprises as well, including insights into how the brain adapts to extended food insufficiency and how migrating cells follow a path through the body. It’s worth looking back on some of the best of that work before the revelations of the coming year give us a new perspective on ourselves again.

Harol Bustos for Quanta Magazine
A Turning Point in Alzheimer’s Research?
Many people connected to Alzheimer’s disease, either through research or through personal ties to patients, hoped that 2022 would be a banner year. Major clinical trials would finally reveal whether two new drugs addressing the perceived root cause of the disease worked. The results fell unfortunately short of expectations. One of the drugs, lecanemab, showed potential for slightly slowing the cognitive decline of some patients but was also linked to sometimes fatal side effects; the other, gantenerumab, was deemed an outright failure.
The disappointing outcomes cap three decades of research based heavily on the theory that Alzheimer’s disease is caused by plaques of amyloid proteins that build up between brain cells and kill them. Mounting evidence suggests, however, that amyloid is only one component in a much more complex disease process that involves damaging inflammation and malfunctions in how cells recycle their proteins. Most of these ideas have been around for as long as the amyloid hypothesis but are only just beginning to receive the attention they deserve.
In fact, aggregations of proteins around cells are beginning to look like an almost universal phenomenon in aging tissues and not a condition peculiar to amyloid and Alzheimer’s disease, according to work by Stanford University researchers that was announced in a preprint last spring. The observation may be one more bit of proof that worsening problems with protein management may be a routine consequence of aging for cells.
Andrey Andreev, Thai Truong, Scott Fraser; Translational Imaging Center, USC
Watching a Memory Form
Neuroscientists have long understood a lot about how memories form — in principle. They’ve known that as the brain perceives, feels and thinks, the neural activity that gives rise to those experiences strengthens the synaptic connections between the neurons involved. Those lasting changes in our neural circuitry become the physical records of our memories, making it possible to re-evoke the electrical patterns of our experiences when they are needed. The exact details of that process have nevertheless been cryptic. Early this year, that changed when researchers at the University of Southern California described a technique for visualizing those changes as they occur in a living brain, which they used to watch a fish learn to associate unpleasant heat with a light cue. To their surprise, while this process strengthened some synapses, it deleted others.
The information content of a memory is only part of what the brain stores. Memories are also encoded with an emotional “valence” that categorizes them as a positive or negative experience. Last summer, researchers reported that levels of a single molecule released by neurons, called neurotensin, seem to act as flags for that labeling.

Samuel Velasco/Quanta Magazine
Molecular Ecosystems Started Life
Life on Earth began with the first appearance of cells roughly 3.8 billion years ago. But paradoxically, before there were cells, there must have been collections of molecules doing surprisingly lifelike things. Over the past decade, researchers in Japan have been conducting experiments with RNA molecules to learn whether a single type of replicating molecule could evolve into a throng of different replicators, as researchers on the origin of life have theorized must have happened in nature. The Japanese scientists found that this diversification did occur, with various molecules coevolving into competing hosts and parasites that rose and fell in dominance. Last March, the scientists reported a new development: The diverse molecules had started working together in a more stable ecosystem . Their work suggests that RNAs and other molecules in the prebiotic world could likewise have coevolved to lay the foundations of cellular life.
Self-replication is often treated as the essential first step in any origin-of-life hypothesis, but it doesn’t have to be. This year, Nick Lane and other evolutionary biologists continued to find evidence that before cells existed, systems of “proto-metabolism” involving complex sets of energetic reactions might have arisen in the porous materials near hydrothermal vents.
Jonas Hartmann/Mayor Lab
How Body Cells Find Their Place
How does a single fertilized egg cell grow into an adult human body with upward of 30 trillion cells in more than 200 specialized categories? It’s the quintessential mystery of development. For much of the past century, the predominant explanation has been that chemical gradients established in various parts of the developing body guide cells to where they’re needed and tell them how to differentiate into the constituents of skin, muscles, bones, brain and other organs.
But chemicals now seem to be only part of the answer. Recent work suggests that while cells do use chemical gradient clues to guide their navigation, they also follow patterns of physical tension in the tissues that surround them, like tightrope walkers crossing a taut cable. Physical tension does more than tell cells where to go. Other work reported in May showed that mechanical forces inside an embryo also help induce sets of cells to become specific structures , such as feathers instead of skin.
Meanwhile, synthetic biologists — researchers who take an engineering approach to the study of life — made important progress in understanding the kinds of genetic algorithms that control how cells differentiate in response to chemical cues. A team at Caltech demonstrated an artificial network of genes that could stably transform stem cells into a number of more specialized cell types. They haven’t identified what the natural genetic control system in cells is, but the success of their model proves that whatever the real system is, it probably doesn’t need to be much more complicated.

Matt Curtis for Quanta Magazine
What the Low-Energy Brain Sees
The brain is the most energy-hungry organ in the body, so perhaps it’s not surprising that evolution devised an emergency strategy to help brains cope with lengthy periods of food deficiency. Researchers at the University of Edinburgh discovered that when mice have to survive on short rations for weeks on end, their brains start to operate in the equivalent of a “low power” mode .
In this state, neurons in the visual cortex use almost 30% less energy at their synapses. From an engineering standpoint, it’s a neat solution for stretching the brain’s energy resources, but there’s a catch. In effect, the low-power mode reduces the resolution of the animal’s vision by making the visual system process signals less precisely.
An engineering view of the brain also recently improved our understanding of another sensory system: our sense of smell. Researchers have been trying to improve the ability of computerized “artificial noses” to recognize smells. Chemical structures alone go a long way toward defining the smells we associate with various molecules. But new work suggests that the metabolic processes that create molecules in nature also reflect our sense of the smell of the molecules. Neural networks that included metabolic information in their analyses came significantly closer to classifying smells the way humans do.
Paşca Lab, Stanford University
Human Brain Cells Connect Inside Animal Brains
A living human brain is still a maddeningly difficult thing for neuroscientists to study: The skull obstructs their view and ethical considerations rule out many potentially informative experiments. That’s why researchers have begun growing isolated brain tissue in the laboratory and letting it form “organoids” with physical and electrical similarities to real brains. This year, the neuroscientist Sergiu Paşca and his colleagues showed how far those similarities go by implanting human brain organoids into newborn laboratory rats. The human cells integrated themselves into the animal’s neural circuitry and took on a role in its sense of smell. Moreover, the transplanted neurons looked healthier than ones growing in isolated organoids, which suggests, as Paşca noted in an interview with Quanta , the importance of providing neurons with inputs and outputs. The work points the way toward developing better experimental models for human brains in the future.
Get highlights of the most important news delivered to your email inbox
Also in Biology
How Our Longest Nerve Orchestrates the Mind-Body Connection
How Colorful Ribbon Diagrams Became the Face of Proteins

The Viral Paleontologist Who Unearths Pathogens’ Deep Histories
Comment on this article.
Quanta Magazine moderates comments to facilitate an informed, substantive, civil conversation. Abusive, profane, self-promotional, misleading, incoherent or off-topic comments will be rejected. Moderators are staffed during regular business hours (New York time) and can only accept comments written in English.

Next article
Use your social network.
Forgot your password ?
We’ll email you instructions to reset your password
Enter your new password
Secondary Menu
- Research Articles & Papers
Korunes, KL; Myers, RB; Hardy, R; Noor, MAF
Drosophila pseudoobscura is a classic model system for the study of evolutionary genetics and genomics. Given this long-standing interest, many genome sequences have accumulated for D. pseudoobscura and closely related species D. persimilis, D. miranda, and D. lowei. To facilitate the exploration… read more about this publication »
Zipple, MN; Roberts, EK; Alberts, SC; Beehner, JC
Bartoš et al. (2021; Mammal Review 51: 143–153; https://doi.org/10.1111/mam.12219) reviewed the mechanisms involved in the ‘Bruce effect’ – a phenomenon originally documented in inseminated female house mice Mus musculus, who block pregnancy following exposure to a novel (non-sire) male. They argue… read more about this publication »
Byrne, M; Koop, D; Strbenac, D; Cisternas, P; Yang, JYH; Davidson, PL; Wray, G
The molecular mechanisms underlying development of the pentameral body of adult echinoderms are poorly understood but are important to solve with respect to evolution of a unique body plan that contrasts with the bilateral body plan of other deuterostomes. As Nodal and BMP2/4 signalling is involved… read more about this publication »
Wang, Q; Xu, P; Sanchez, S; Duran, P; Andreazza, F; Isaacs, R; Dong, K
BackgroundInsects rely on their sense of smell to locate food and hosts, find mates and select sites for laying eggs. Use of volatile compounds, such as essential oils (EOs), to repel insect pests and disrupt their olfaction-driven behaviors has great practical significance in integrated pest… read more about this publication »
Castano-Duque, L; Ghosal, S; Quilloy, FA; Mitchell-Olds, T; Dixit, S
Rice production is shifting from transplanting seedlings to direct sowing of seeds. Following heavy rains, directly sown seeds may need to germinate under anaerobic environments, but most rice (Oryza sativa) genotypes cannot survive these conditions. To identify the genetic architecture of complex… read more about this publication »
Peng, L; Shan, X; Wang, Y; Martin, F; Vilgalys, R; Yuan, Z
Clitopilus hobsonii (Entolomataceae, Agaricales, Basidiomycetes) is a common soil saprotroph. There is also evidence that C. hobsonii can act as a root endophyte benefitting tree growth. Here, we report the genome assembly of C. hobsonii QYL-10, isolated from ectomycorrhizal root tips of Quercus… read more about this publication »
Yan, W; Wang, B; Chan, E; Mitchell-Olds, T
The genetic basis of flowering time changes across environments, and pleiotropy may limit adaptive evolution of populations in response to local conditions. However, little information is known about how genetic architecture changes among environments. We used genome-wide association studies (GWAS… read more about this publication »
Doak, DF; Waddle, E; Langendorf, RE; Louthan, AM; Isabelle Chardon, N; Dibner, RR; Keinath, DA; Lombardi, E; Steenbock, C; Shriver, RK; Linares, C; Begoña Garcia, M; Funk, WC; Fitzpatrick, SW; Morris, WF; Peterson, ML
Structured demographic models are among the most common and useful tools in population biology. However, the introduction of integral projection models (IPMs) has caused a profound shift in the way many demographic models are conceptualized. Some researchers have argued that IPMs, by explicitly… read more about this publication »
Kim, JH; Hilleary, R; Seroka, A; He, SY
A grand challenge facing plant scientists today is to find innovative solutions to increase global crop production in the context of an increasingly warming climate. A major roadblock to global food sufficiency is persistent loss of crops to plant diseases and insect infestations. The United… read more about this publication »
Yuan, M; Jiang, Z; Bi, G; Nomura, K; Liu, M; Wang, Y; Cai, B; Zhou, J-M; He, SY; Xin, X-F
The plant immune system is fundamental for plant survival in natural ecosystems and for productivity in crop fields. Substantial evidence supports the prevailing notion that plants possess a two-tiered innate immune system, called pattern-triggered immunity (PTI) and effector-triggered immunity (… read more about this publication »
Benito-Kwiecinski, S; Giandomenico, SL; Sutcliffe, M; Riis, ES; Freire-Pritchett, P; Kelava, I; Wunderlich, S; Martin, U; Wray, GA; McDole, K; Lancaster, MA
The human brain has undergone rapid expansion since humans diverged from other great apes, but the mechanism of this human-specific enlargement is still unknown. Here, we use cerebral organoids derived from human, gorilla, and chimpanzee cells to study developmental mechanisms driving evolutionary… read more about this publication »
Lofgren, LA; Nguyen, NH; Vilgalys, R; Ruytinx, J; Liao, H-L; Branco, S; Kuo, A; LaButti, K; Lipzen, A; Andreopoulos, W; Pangilinan, J; Riley, R; Hundley, H; Na, H; Barry, K; Grigoriev, IV; Stajich, JE; Kennedy, PG
While there has been significant progress characterizing the 'symbiotic toolkit' of ectomycorrhizal (ECM) fungi, how host specificity may be encoded into ECM fungal genomes remains poorly understood. We conducted a comparative genomic analysis of ECM fungal host specialists and generalists,… read more about this publication »
Mitchell, RM; Ames, GM; Wright, JP
Background and aimsUnderstanding impacts of altered disturbance regimes on community structure and function is a key goal for community ecology. Functional traits link species composition to ecosystem functioning. Changes in the distribution of functional traits at community scales in response to… read more about this publication »
DeMarche, ML; Bailes, G; Hendricks, LB; Pfeifer-Meister, L; Reed, PB; Bridgham, SD; Johnson, BR; Shriver, R; Waddle, E; Wroton, H; Doak, DF; Roy, BA; Morris, WF
Spatial gradients in population growth, such as across latitudinal or elevational gradients, are often assumed to primarily be driven by variation in climate, and are frequently used to infer species' responses to climate change. Here, we use a novel demographic, mixed-model approach to dissect the… read more about this publication »
Shaw, EC; Fowler, R; Ohadi, S; Bayly, MJ; Barrett, RA; Tibbits, J; Strand, A; Willis, CG; Donohue, K; Robeck, P; Cousens, RD
Aim: If we are able to determine the geographic origin of an invasion, as well as its known area of introduction, we can better appreciate the innate environmental tolerance of a species and the strength of selection for adaptation that colonizing populations have undergone. It also enables us to… read more about this publication »
Rushworth, CA; Mitchell-Olds, T
Despite decades of research, the evolution of sex remains an enigma in evolutionary biology. Typically, research addresses the costs of sex and asexuality to characterize the circumstances favoring one reproductive mode. Surprisingly few studies address the influence of common traits that are, in… read more about this publication »
Jorge, JF; Bergbreiter, S; Patek, SN
Small organisms can produce powerful, sub-millisecond impacts by moving tiny structures at high accelerations. We developed and validated a pendulum device to measure the impact energetics of microgram-sized trap-jaw ant mandibles accelerated against targets at 105 m s-2 Trap-jaw ants (… read more about this publication »
Markunas, AM; Manivannan, PKR; Ezekian, JE; Agarwal, A; Eisner, W; Alsina, K; Allen, HD; Wray, GA; Kim, JJ; Wehrens, XHT; Landstrom, AP
Long QT syndrome (LQTS) is a genetic disease resulting in a prolonged QT interval on a resting electrocardiogram, predisposing affected individuals to polymorphic ventricular tachycardia and sudden death. Although a number of genes have been implicated in this disease, nearly one in four… read more about this publication »
Stone, DF; Mccune, B; Pardo-De La Hoz, CJ; Magain, N; Miadlikowska, J
The new genus Sinuicella, an early successional lichen, was found on bare soil in Oregon, USA. The thallus is minute fruticose, grey to nearly black, branching isotomic dichotomous, branches round, 20-90 μm wide in water mount. The cortex is composed of interlocking cells shaped like jigsaw puzzle… read more about this publication »
Oita, S; Ibáñez, A; Lutzoni, F; Miadlikowska, J; Geml, J; Lewis, LA; Hom, EFY; Carbone, I; U'Ren, JM; Arnold, AE
Understanding how species-rich communities persist is a foundational question in ecology. In tropical forests, tree diversity is structured by edaphic factors, climate, and biotic interactions, with seasonality playing an essential role at landscape scales: wetter and less seasonal forests… read more about this publication »
Hibshman, JD; Webster, AK; Baugh, LR
Standard laboratory culture of Caenorhabditis elegans utilizes solid growth media with a bacterial food source. However, this culture method limits control of food availability and worm population density, factors that impact many life-history traits. Here, we describe liquid-culture protocols for… read more about this publication »
Caves, EM; Green, PA; Zipple, MN; Bharath, D; Peters, S; Johnsen, S; Nowicki, S
AbstractSensory systems are predicted to be adapted to the perception of important stimuli, such as signals used in communication. Prior work has shown that female zebra finches perceive the carotenoid-based orange-red coloration of male beaks-a mate choice signal-categorically. Specifically,… read more about this publication »
Reed, PB; Peterson, ML; Pfeifer-Meister, LE; Morris, WF; Doak, DF; Roy, BA; Johnson, BR; Bailes, GT; Nelson, AA; Bridgham, SD
Predicting species' range shifts under future climate is a central goal of conservation ecology. Studying populations within and beyond multiple species' current ranges can help identify whether demographic responses to climate change exhibit directionality, indicative of range shifts, and whether… read more about this publication »
Sallee, JL; Crawford, JM; Singh, V; Kiehart, DP
Actin filament crosslinking, bundling and molecular motor proteins are necessary for the assembly of epithelial projections such as microvilli, stereocilia, hairs, and bristles. Mutations in such proteins cause defects in the shape, structure, and function of these actin - based protrusions. One… read more about this publication »
- Duke Biology’s Mission Statement
- AJED Annual and Semester Reports
- AJED Meeting Notes
- Biology Cultural Association (BCA)
- Inclusion, Diversity, Equity, and Antiracism Committee (IDEA)
- Learning from Baboons: Dr. Susan Alberts
- Extremophiles and Systems Biology: Dr. Amy Schmid
- How Cells Manage Stress: Dr. Gustavo Silva
- Predator-Prey Interactions in a Changing World: Dr. Jean Philippe Gibert
- Exploring the Extracellular Matrix: Dr. David Sherwood
- Cell Division's Missing Link: Dr. Masayuki Onishi
- Listening in to Birdsong: Dr. Steve Nowicki
- Biogeochemistry as Ecosystem Accounting: Dr. Emily Bernhardt
- Building a Dynamic Nervous System: Dr. Pelin Volkan
- Investigating a Key Plant Hormone: Dr. Lucia Strader
- Imagining Visual Ecology: Dr. Sönke Johnsen
- Outreach Opportunities Across the Triangle
- Job Opportunities
- Location & Contact
- Frequently Asked Questions
- Learning Outcomes
- Major Requirements
- Anatomy, Physiology & Biomechanics
- Animal Behavior
- Biochemistry
- Cell & Molecular Biology
- Evolutionary Biology
- Marine Biology
- Neurobiology
- Pharmacology
- Plant Biology
- Minor Requirements
- Biology IDM
- List of Biology Advisors
- Guide for First-Year Students
- Transfer Credit
- Application & Deadlines
- Supervisor & Faculty Reader
- Thesis Guidelines
- Honors Poster
- Past Student Projects
- Study Away Opportunities
- Finding a Research Mentor
- Project Guidelines
- Getting Registered
- Writing Intensive Study
- Independent Study Abroad
- Summer Opportunities
- Departmental Awards
- Biology Majors Union
- Commencement 2024
- Trinity Ambassadors
- Degree Programs
- Ph.D. Requirements
- How to Apply
- Financial Aid
- Living in Durham
- Where Our Students Go
- Milestones Toward Ph.D.
- Graduate School Fellowships
- Useful Resources
- Concurrent Biology Master of Science
- En Route Biology Masters of Science
- Form Library
- Mentorship Expectations
- On Campus Resources
- Fellowships & Jobs
- Meet Our Postdocs
- Department Research Areas
- Research Facilities
- Duke Postdoctoral Association
- All Courses
- Biological Structure & Function Courses
- Ecology Courses
- Organismal Diversity Courses
- Alternate Elective Courses
- Primary Faculty
- Secondary Faculty
- Graduate Faculty
- Emeritus Faculty
- Graduate Students
- Department Staff
- Faculty Research Labs
- Developmental Biology
- Ecology & Population Biology
- Neuroscience
- Organismal Biology & Behavior
- Systematics
- Botany Plot
- Field Station
- Pest Management Protocols
- Research Greenhouses
- Centers/Research Groups
- Biology Writes
- Alumni Profiles
- For Current Students
- Assisting Duke Students
- Election 2024
- Entertainment
- Newsletters
- Photography
- AP Buyline Personal Finance
- AP Buyline Shopping
- Press Releases
- Israel-Hamas War
- Russia-Ukraine War
- Global elections
- Asia Pacific
- Latin America
- Middle East
- Election results
- Google trends
- AP & Elections
- U.S. Open Tennis
- Paralympic Games
- College football
- Auto Racing
- Movie reviews
- Book reviews
- Financial Markets
- Business Highlights
- Financial wellness
- Artificial Intelligence
- Social Media
Mushrooms foraged in Sweden could help research Chernobyl fallout
A golden chanterelle mushroom, shown here in Stockholm, Sweden, on July 31, 2021. (AP Photo/Natalie Li)
FILE - A shelter construction covers the exploded reactor at the Chernobyl nuclear plant, in Chernobyl, Ukraine, April 27, 2021. (AP Photo/Efrem Lukatsky, File)
- Copy Link copied
COPENHAGEN, Denmark (AP) — Sweden’s strong foraging culture could help determine how much radioactive fallout remains in the Scandinavian country 38 years after the Chernobyl nuclear explosion .
The Swedish Radiation Safety Authority has asked mushroom-pickers to send samples of this season’s harvest for testing. The goal of the measurement project is to map the levels of Cesium-137 in mushrooms, which can absorb the isotope from soil, and see how much remains after the April 26, 1986 disaster at the Soviet nuclear power plant in what is now Ukraine.
Cesium, the key radioactive material released in the fallout, has a half-life of some 30 years. It can build up in the body, and high levels are thought to be a risk.
The radiation watchdog is counting on the foraging lifestyle in Sweden, which is covered by more than 60% of forest, to aid its research. In late summer, many Swedes spend days in the woods collecting berries, mushrooms and plants.
It’s asking foragers where they found their bounty — though they don’t have to disclose the exact whereabouts of the prized golden chanterelle mushroom.
Spots that regularly produce such chanterelles — often called “the gold of the forest mushroom” — are closely guarded family secrets that could cause headaches for researchers who need data points.
“It doesn’t have to be the exact location of the most secret chanterelle spot,” said Pål Andersson, an investigator at the Radiation Safety Authority.
Mushroom-pickers are instructed to send in double-bagged edible fungi — at least 100 grams (3.53 ounces) of fresh mushrooms, or 20 grams (0.71 ounces) of dried mushrooms — picked in 2024.
Sweden’s safety authority did not say when a result of its research was expected.
Dozens of people were killed in the immediate aftermath of the Chernobyl disaster, while the radioactive fallout spread across Europe. The long-term death toll from radiation poisoning is unknown.
Swedish authorities were the first to detect radioactive fallout in Europe, forcing Soviet officials, who had attempted to cover up the disaster, to open up about it days later.
In 2017, a state veterinary agency in the Czech Republic said about half of all wild boars in the country’s southwest were radioactive and considered unsafe for consumption. The boars feed on an underground mushroom that absorbs radioactivity from the soil. Similar problems with radioactive wild animals were reported in Austria and Germany.
Dazio reported from Berlin.
What We Publish
Plos biology article types.
PLOS Biology considers works of exceptional significance, originality, and relevance in all areas of biological science, including both primary research, meta-analyses and Magazine articles . Our publication options are outlined below.
Research-based content
PLOS Biology publishes seven different types of research reports. All research articles are compatible with our easy, format-free submission process, and offer options for preprints, published peer review history, and publishing uncorrected proofs. Most, with rare exception, are also protected by our scooping policy , ensuring that your research will not be rejected for novelty within six-months of the publication of a complementary or confirmatory research publication. We evaluate all research based on the important questions it answers and its potential to impact an international scientific community as well as educators, policy makers, patient advocacy groups, and society more broadly.
Research Articles
|
| Research Articles are the backbone of and the type of research we publish most frequently. We publish high-caliber research of any length, spanning the full breadth of the biological sciences, from molecules to ecosystems. We also consider works at the interface of other disciplines, including research of interest to the clinical and pre-clinical research communities. To be appropriate for , translational biological research should demonstrate the potential to advance our insights into the understanding, detection, diagnosis, prevention, and/or treatment of human disease. |
Preregistered Research Articles
|
| Preregistered Research Articles (also known as Registered Reports) are a form of empirical research in which the study design and proposed analyses are peer reviewed prior to conducting experiments, data collection or analysis. Studies addressing important research questions are reviewed for technical soundness of the proposed methodology, and provisionally accepted for publication before data collection commences. This approach is designed to minimize publication and research bias while also maximizing study quality by focusing peer review on the importance of the research question and rigour of the proposed methodology. It also allows complete flexibility to conduct exploratory analyses and report serendipitous findings. Submission and assessment takes place in two stages before and after the investigation, and results in a single cumulative publication. |
Methods & Resources Articles
|
| Methods and Resources Articles describe technical innovations, including novel approaches to a previously inaccessible biological innovation, or substantial improvements over previously established methods. The reported method should be thoroughly validated, and while presenting new biological insights is encouraged, this is not a requirement for consideration. Resources consist of data sets or other significant scientific resources that are of general interest and provide exceptionally value for the community that could spur future research. |
Meta-Research Articles
|
| Meta-Research Articles examine how biological research is designed, carried out, communicated and evaluated, or explore the systems that evaluate and reward individual scientists or institutions in new and novel ways. We welcome both exploratory and confirmatory research that has the potential to drive change in research and evaluation practices in the life sciences and beyond. Themes include, but are not limited to, transparency, established and novel methodological standards, sources of bias (conflicts of interest, selection, inflation, funding, etc.), data sharing, evaluation metrics, assessment, reward, and funding structures. Meta-research articles are not meant for meta-analyses of biological data (please submit these as Research Articles). |
Short Reports
|
| Short Reports are tailored for concise communication of impactful discoveries. These reports offer a self-contained platform to share brief yet compelling research findings. They present results from a limited set of experiments, typically summarized in 3-4 figures or fewer. The outcomes should be self-contained, rather than fitting within the narrative arc of a larger research project or article. Short Reports should be organized as the . |
Discovery Reports
|
| Discovery Reports describe novel and intriguing initial findings with the potential to lead to a significant new result for the field. Discovery Reports are short articles, typically with 2-4 main figures. While the research may be preliminary, studies should be advanced to the stage where observations or findings have been confirmed by independent methods or experimental approaches and obvious alternative interpretations have been ruled out. Discovery Reports are designed to work together with Update Articles to empower researchers to evaluate and share work in a way that more closely mirrors the real-world research process and create a comprehensive research story. |
| Like all ’s articles, Discovery Reports are assessed on the basis of significance, originality, and relevance to biological science, with an additional focus on the value and interest of the research question posed. | |
| Discovery Reports undergo the same rigorous as other articles. Reviewers will focus particularly on the robustness and validity of the result to ensure the reported findings are not artifacts or false positives. | |
| will not apply if a published paper has already elucidated the mechanism/phenomenon being reported but the Discovery Report has not. |
Update Articles
|
| Update Articles develop a previous PLOS Biology study by providing new, robust mechanistic insight, identifying the biological or physiological significance of the previous findings, or in another way significantly adding to the original article. Results should go well beyond confirming the original observations. Recognizing the importance of correcting the scientific record when needed, we do consider negative Update Articles. Negative updates require clear, well-supported experiments demonstrating and explaining why the initial observation did not work as expected, or did not hold up to further scrutiny. Negative updates should stand alone as a research article; shorter contributions contesting specific aspects of the execution or analysis of a PLOS Biology article may be more appropriate as a . |
| Like all ’s articles, Update Articles are assessed on the basis of originality and relevance to biological science. A high level of rigour is expected in , execution, and substantial evidence for conclusions. | |
| Update Articles undergo the same rigorous as other articles. We strive to consult the same Academic Editor who evaluated the original study and 1-2 of the same reviewers, if appropriate considering the time elapsed from the original publication. New reviewers may be needed to cover all areas of expertise relevant to the follow up experiments in the Update Article. Depending on the circumstances, in cases where a different lab has submitted an update, the authors of the original research may be invited to provide signed comments. |
Magazine articles
Our magazine section features non-research articles that cover topical issues and are accessible to a broad audience while remaining scientifically rigorous. Magazine readers include scientists, scientific educators, students, physicians, patients, and the interested public. There is no publication fee for magazine articles.
Our magazine section is divided into Front Matter and In-depth Analysis subsections. Front Matter articles are short, focussed and provide opinion on topical issues, community resources or commentary on PLOS Biology articles. In-depth Analyses are long-form articles providing forward-looking analysis of a given topic, highlighting gaps in our current understanding or putting forward community recommendations or guidelines. We do not publish traditional review articles.
Most magazine articles are comissioned by the editors, but we do publish some unsolicted content. If you have a suggestion for any of our current magazine categories, we will consider the idea. Before finalizing the piece, we recommend that you discuss it with us by sending a presubmission enquiry via email to [email protected] . Before submitting, consult our General Guidelines for Magazine Submissions , as well as the guidelines for the specific article type you are interested in.
If you have any questions about submitting an idea or article for consideration in the magazine section, email our editorial team at [email protected] .
General Guidelines for Magazine Submissions
Magazine content is intended for the broader biological community including students, scientists, and the educated general reader. It is therefore important that the writing style be concise, clear and accessible. Avoid specialist terms, abbreviations and jargon. Editors will make suggestions to make your piece more accessible, as well as cuts or additions that could strengthen the article. Our aim is to make the editorial process rigorous and consistent, but not intrusive or overbearing.
If your manuscript includes original research data/analyses, you must ensure that it complies with PLOS’ data policy . If this is the case, it is likely that the submission would be more appropriate for one of our research article types. Please see Research-based content for more information.
PLOS applies the Creative Commons Attribution (CC BY) license to all figures we publish, which allows them to be freely used, distributed, and built upon as long as proper attribution is given. Read more about our content license . DO NOT submit any figures that have been previously copyrighted or contain proprietary data unless you have and can supply written permission from the copyright holder to use that content. If in doubt, contact our editorial office .
Funding Statement
As part of the PLOS Biology submission form you’ll be asked to provide a funding statement, which will be published with the article if the manuscript is accepted. Your funding statement should describe any funding that helped to support the work, as follows:
- Include grant numbers and the URLs of any funder's website. Use the full name, not acronyms, of funding institutions, and use initials to identify authors who received the funding.
- Describe the role of any sponsors or funders in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. If the funders had no role in any of the above, include this sentence at the end of your statement: "The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript."
- If the study was unfunded , please provide the following statement: "The author(s) received no specific funding for this work."
How to Submit
Follow the main submission guidelines , and consult our figures , tables , and supporting information pages when preparing your manuscript.
Submit your manuscript through our submission system . When you start your submission, select the appropriate article type from the dropdown menu. Manuscripts can be submitted as DOC, DOCX, or PDF files.
Front Matter
Editorials are written in-house by members of the editorial staff or by members of the Editorial Board.
Perspectives
The Perspectives section provides experts with a forum to comment on topical or controversial issues of broad interest. They address controversial issues or those at the interface between science and policy or science and society; present a policy position aimed at influencing policy decisions; examine and make recommendations on scientific and publishing practices. These are meant to be short, opinionated, Op-ed type of pieces.
The ideal Perspective conveys a sense of urgency. Some things to think about would be:
- Is this topic of immediate concern?
- Is the topic relevant to a pressing regional or global issue
- Does the piece offer a novel point of view on a scientific or policy issue, or on topical events?
- Does is make specific, practical proposals to address the issue?
Controversial articles are welcomed, but the text should acknowledge that a position is in fact controversial and provide readers with enough background on the differing views.
Our Perspectives aim to engage a broad and diverse audience—it is therefore important to ensure that they are written in an accessible, persuasive, and stimulating style that appeals to both specialists and non-specialist readers. Perspectives are usually assessed in-house with our Editorial Board, but we reserve the right to peer-review them if needed. Commissioning does not guarantee publication. Editors work closely with authors to ensure that articles are written in an engaging, succinct, yet rigorous manner.
Guidelines for a Perspective
| Title length | up to 75 characters |
| Standfirst length | up to 260 characters |
| Manuscript length | ~1,000 |
| References | 10 |
| Display items (figures, text boxes, tables) | 1 if needed, but not necessary Submitted figures must be publishable under the , thus, with few exceptions, we cannot accept previously published work |
Example Perspectives
Lowe-Power T, Dyson L, Polter AM (2021) A generation of junior faculty is at risk from the impacts of COVID-19. PLoS Biol 19(5): e3001266. https://doi.org/10.1371/journal.pbio.3001266
Bourne PE (2021) Is “bioinformatics” dead? PLoS Biol 19(3): e3001165. https://doi.org/10.1371/journal.pbio.3001165
Primers provide concise and accessible context to a PLOS Biology research article of broad and current interest. Primers are commissioned and published alongside a research article that would benefit from additional context and/or explanation.
Above all, Primers should demystify an area of biology, avoid and/or explain technical jargon and provide critical and forward-thinking analysis about how the research article fits into the current state of the field and its future. A good Primer will briefly discuss (but not exhaustively review) what we know and what questions we have yet to answer for a particular field. It will then introduce the new findings and describe in roughly three paragraphs the advance represented in the related research article, highlighting its significance, not only for the discipline in question, but across disciplines. The Primer should then explain what the findings suggest in terms of next steps: what new avenues of investigation are opened, what new experiments can be tried, what new ideas can now be tested going forward? Ideally, Primers also offer insight into what big questions are likely to remain unanswered for many years (for whatever reasons).
We encourage the use of a figure to illustrate key concepts/mechanism/conclusions in an informative, easy-to-understand manner.
Primers are usually assessed by our Editorial Board, but we reserve the right to peer-review externally if needed. Commissioning does not guarantee publication. Editors work closely with authors to ensure that articles are written in an engaging, succinct, yet rigorous manner.
Guidelines for a Primer
| Title length | up to 75 characters |
| Standfirst length | up to 260 characters |
| Manuscript length | ~1,000 words |
| References | 10 |
| Display items (figures, text boxes, tables) | 1 Submitted figures must be publishable under the , thus, with few exceptions, we cannot accept previously published work |
Example Primer
Kazanova A, Rudd CE (2021) Programmed cell death 1 ligand (PD-L1) on T cells generates Treg suppression from memory. PLoS Biol 19(5): e3001272. https://doi.org/10.1371/journal.pbio.3001272
Guizetti J, Frischknecht F (2021) Apicomplexans: A conoid ring unites them all. PLoS Biol 19(3): e3001105. https://doi.org/10.1371/journal.pbio.3001105
Community Pages
Community Pages provide individuals, networks and organizations with the opportunity to highlight resources, tools, or initiatives of benefit to the scientific community and beyond (including science education and public engagement in science). All resources or tools, and the outputs of initiatives must be open and accessible to all.
Contributors must resist the temptation of self-promotion and instead focus on conveying information to a diverse audience.Community Pages should be written in a succinct, accessible, semi-journalistic style that captures the interest of both specialists and non-specialist readers. We encourage the use of 1-2 figures to illustrate key concepts in an informative, easy to grasp manner; or the use of text boxes for background, self-contained information.
Community Pages are peer-reviewed and commissioning does not guarantee publication. Editors work closely with authors to ensure that articles are written in an engaging, succinct, yet rigorous manner.
Guidelines for a Community Page
| Title length | up to 75 characters |
| Standfirst Abstract length | up to 260 characters |
| Manuscript length | 1,000-1,200 words |
| References | 10 |
| Display items (figures, text boxes, tables) | 1-2 Submitted figures must be publishable under the , thus, with few exceptions, we cannot accept previously published work |
Example Community Pages
Weissgerber TL (2021) Training early career researchers to use meta-research to improve science: A participant-guided “learn by doing” approach. PLoS Biol 19(2): e3001073. https://doi.org/10.1371/journal.pbio.3001073
McCullagh EA, Nowak K, Pogoriler A, Metcalf JL, Zaringhalam M, Zelikova TJ (2019) Request a woman scientist: A database for diversifying the public face of science. PLoS Biol 17 (4): e3000212. https://doi.org/10.1371/journal. pbio.3000212
Formal Comments
Formal Comments are intended to provide a formal outlet for the discussion and interpretation of research findings associated with specific articles published in PLOS Biology . They are designed to ensure that readers obtain a balanced view of a scientific or meta-scientific/policy question, especially in areas of debate/controversy. Formal Comments are peer-reviewed and indexed in PubMed.
Formal Comments must be coherent, concise, and well-argued, and are subject to the PLOS Biology criteria for publication . Editors will, as a matter of course, invite the authors of the original article to submit a response to the Formal Comment. Any revisions (of the Formal Comment or the response) will be shared with the authors of the associated comment.
Guidelines for Formal Comments
| Title length | up to 75 characters |
| Manuscript length | Formal Comments:1,000 words |
| References | ~10 |
| Display items (figures, text boxes, tables) | 1-2 Submitted figures must be publishable under the , thus, with few exceptions, we cannot accept previously published work |
Example Formal Comment
Rees WE, Wackernagel M (2013) The Shoe Fits, but the Footprint is Larger than Earth. PLoS Biol 11(11): e1001701. https://doi.org/10.1371/journal.pbio.1001701
Example Formal Comment - Response
Blomqvist L, Brook BW, Ellis EC, Kareiva PM, Nordhaus T, Shellenberger M (2013) The Ecological Footprint Remains a Misleading Metric of Global Sustainability. PLoS Biol 11(11): e1001702. https://doi.org/10.1371/journal.pbio.1001702
In-Depth Analysis
Essays are opinionated articles on a topic of interest to scientists and to a broader audience, including the general public. Unlike traditional review articles, which include a comprehensive account of a field, Essays take an imaginative approach to a provocative question, with an engaging but rigorous investigation of the problem. We encourage authors of Essays to select the most representative references to convey their points and avoid exhaustively covering the relevant literature.
The remit of Essays is very broad. They might:
- take stock of progress in a field from a personal point of view
- explore the implications of recent advances that promise to have broad-ranging consequences on a field
- comment on a topical or controversial area of research
- discuss key ideas or educational strategies to enhance understanding of fundamental biological questions
- offer historical/philosophical reflections on contemporary biology
- analyze scientific issues with policy implications
Our Essays aim to engage a broad and diverse audience—it is therefore important to ensure that they are written in an accessible, semi-journalistic style that captures the interest of both specialists and non-specialist readers. We encourage the use of figures to illustrate key concepts in an informative, easy to grasp manner; as well as the use of text boxes for background, self-contained information.
Essays are peer-reviewed and commissioning does not guarantee publication.
Guidelines for an Essay
| Title length | up to 75 characters |
| Abstract length | ~100 words |
| Manuscript length | ~3,500-4,000 words
|
| References | ~30-60
|
| Display items (figures, text boxes, tables) | up to 3-4 Submitted figures must be publishable under the , thus, with few exceptions, we cannot accept previously published work |
Example Essays
Rees T, Bosch T, Douglas AE (2018) How the microbiome challenges our concept of self. PLoS Biol 16(2): e2005358. https://doi.org/10.1371/journal.pbio.2005358 Konig C, Weigelt P, Schrader J, Taylor A, Kattge J, Kreft H (2019) Biodiversity data integration—the significance of data resolution and domain. PLoS Biol 17(3): e3000183. https://doi.org/10.1371/journal.pbio.3000183
Unsolved Mysteries
Unsolved Mysteries discuss a topic of biological or medical importance that is poorly understood and in need of research attention—e.g., an unexplored or challenging question, an emerging opportunity, or a recent puzzling phenomenon. The articles are intended to stimulate the scientific community to think about future research possibilities outside their areas of expertise. The articles should be aimed at a very broad audience of biologists—an unsolved mystery in a neuroscience topic should be accessible to ecologists and biophysicists, for example. The article should include a discussion of the basic science relevant to the topic, why it is biologically or medically important, what work has been done on the topic (if any), major challenges to understanding the question at hand, competing hypotheses, and what advances would be necessary to shed light on the problem. Ideally the structure of the article should reflect the mystery (e.g. subsections with questions as headings). The article should end with a discussion of possible means to a solution
Unsolved Mysteries are peer-reviewed and commissioning does not guarantee publication. Editors work closely with authors to ensure that articles are written in an engaging, succinct, yet rigorous manner.
Guidelines for an Unsolved Mystery
| Title length | up to 75 characters |
| Abstract length | ~100 words |
| Manuscript length | ~3,500-4,000 words
|
| References | ~30-60
|
| Display items (figures, text boxes, tables) | up to 3-4 Submitted figures must be publishable under the , thus, with few exceptions, we cannot accept previously published work |
Example Unsolved Mysteries
Margolis L, Sadovsky Y (2019) The biology of extracellular vesicles: The known unknowns. PLoS Biol 17(7): e3000363. https://doi.org/10.1371/journal.pbio.3000363
Vogels CBF, Ru¨ckert C, Cavany SM, Perkins TA, Ebel GD, Grubaugh ND (2019) Arbovirus coinfection and co-transmission: A neglected public health concern? PLoS Biol 17(1): e3000130. https://doi.org/10.1371/journal.pbio.3000130
Consensus View
Consensus View articles present a comprehensive analysis by an independent and usually multidisciplinary panel of experts who make specific recommendations on important scientific, publishing or policy issues.
Consensus Views are peer-reviewed and commissioning does not guarantee publication. Editors work closely with authors to ensure that articles are written in an engaging, succinct, yet rigorous manner.
Guidelines for a Consensus View
| Title length | up to 75 characters |
| Abstract length | ~100 words |
| Manuscript length | ~4,000-5,000 words
|
| References | There are no strict reference limits, but in general Consensus Views are meant to discuss representative references only |
| Display items (figures, text boxes, tables) | up to 3-4 Submitted figures must be publishable under the , thus, with few exceptions, we cannot accept previously published work |
Example Consensus View
Brown TM, Brainard GC, Cajochen C, Czeisler CA, Hanifin JP, Lockley SW, et al. (2022) Recommendations for daytime, evening, and nighttime indoor light exposure to best support physiology, sleep, and wakefulness in healthy adults. PLoS Biol 20(3): e3001571. https://doi.org/10.1371/journal.pbio.3001571
Kent BA, Holman C, Amoako E, Antonietti A, Azam JM, Ballhausen H, et al. (2022) Recommendations for empowering early career researchers to improve research culture and practice. PLoS Biol 20(7): e3001680. https://doi.org/10.1371/journal.pbio.3001680
Vimercati G, Probert AF, Volery L, Bernardo-Madrid R, Bertolino S, Céspedes V, et al. (2022) The EICAT+ framework enables classification of positive impacts of alien taxa on native biodiversity. PLoS Biol 20(8): e3001729. https://doi.org/10.1371/journal.pbio.3001729
Retired Article Types
Book Review/Science in the Media. These short reviews critiqued books, films, plays, and other media that deal with some aspect of the biological sciences.
- Education. Although we no longer publish Education articles as a separate article type, we continue publishing them as part of the Education Series. The format selected (Essay, Perspective, or Community Page) depends on the aims of each article.
- Historical and Philosophical Perspectives. The Historical and Philosophical Perspectives section provided professional historians and philosophers of science with a forum to reflect on topical issues in contemporary biology.
- Obituaries.
- Open Highlights. Written in-house by members of the editorial staff, Open Highlights used recent publication as keystones around which to nucleate a short synthesis of several related research articles from PLOS and the wider Open Access corpus.
- Research Matters. Brief pieces by leading scientists explaining why the research carried out in their laboratories - and those of their collaborators and their colleagues - matters to lay audiences.
- Series. Series were recurrent themed articles on specific topics, including Education, Public Engagement with Science, Cool Tools, and Where Next?
- Synopses. Selected PLOS Biology research articles are accompanied by a synopsis written for a general audience to provide non-experts with insight into the significance of the published work. They are commissioned only.
Post-Publication Notices
PLOS publishes Editorial Notes, Corrections, Expressions of Concern, and Retraction notices, as needed, to address issues that arise after a PLOS article has been published.

Bhaskaran Publishes Research on Laryngeal Dystonia
Written by Staff
September 3, 2024
Divya Bhaskaran , Assistant Professor in the Exercise Science program of the Biology Department, published a research paper in the Frontiers in Neurology Journal. The article, entitled "Effects of an 11-week vibro-tactile stimulation treatment on voice symptoms in laryngeal dystonia," is a longitudinal clinical trial conducted during Dr Bhaskaran's post-doctoral work at the University of Minnesota.
1536 Hewitt Ave
Saint Paul, MN 55104
General Information
Undergraduate Admission
Public Safety Office
Graduate Admission
ITS Central Service Desk
© 2024 Hamline University
In association with Mitchell | Hamline School of Law ®. Mitchell Hamline School of Law ® has more graduate enrollment options than any other law school in the nation.
- On-Campus Transfer
- Online Degree Completion
- International
- Admitted Students
- How to Apply
- Grants & Scholarships
- First-Year and Transfer Aid
- Online Degree Completion Aid
- Graduate Aid
- International Aid
- Military & Veteran Aid
- Undergraduate Tuition
- Online Degree Completion Tuition
- Graduate Tuition
- Housing & Food Costs
- Net Price Calculator
- Payment Info
- Undergraduate
- Continuing Education
- Program Finder
- Faculty by Program
- College of Liberal Arts
- School of Business
- School of Education & Leadership
- Mitchell Hamline School of Law
- Academic Bulletin
- Academic Calendars
- Bush Library
- Registration & Records
- Study Away & Study Abroad
- Housing & Dining
- Counseling & Health
- Service, Spiritual Life, & Recreation
- Activities & Organizations
- Diversity Resources
- Arts at Hamline
- Meet Our Students
- The Neighborhood
- The Hamline Academic Experience
- Student Research Opportunities
- Paid Internships
- Career Development Center
- Alumni Success Stories
- Center for Academic Success & Achievement
- Writing Center
- Why Hamline?
- Mission & History
- Fast Facts and Rankings
- University Leadership
- Diversity, Equity & Inclusion
- Alumni and Donors
- Request Info
Health | New treatment could be “game-changing tool” in…
Share this:.
- Click to share on Facebook (Opens in new window)
- Click to share on Reddit (Opens in new window)
- Click to share on Twitter (Opens in new window)
Digital Replica Edition
- Latest Headlines
- Environment
- Transportation
- News Obituaries
Health | New treatment could be “game-changing tool” in fight against Alzheimer’s, CSU research finds
Combination of drugs targets two brain proteins critical in neuroinflammation involved in brain aging and alzheimer’s.

The combination of drugs targets two brain proteins critical in neuroinflammation, which is involved in brain aging and Alzheimer’s, according to a study published in July in the Journal of Neuroinflammation featuring CSU researchers.
Results from the study show this medicine could become “a game-changing tool” against Alzheimer’s, researchers said.
“There are no effective treatments right now,” said Devin Wahl, a CSU postdoctoral fellow, who co-authored the study. “We have treatments that can manage symptoms, but we don’t have any that can stop the disease. We want to try to identify novel treatments that may be effective to slow, or even reduce, the effects of Alzheimer’s disease.”
This cocktail of medicines could also improve memory in aging adults, the study found, and, potentially, reverse cognitive decline.
The research came out of a partnership between CSU faculty member Tom LaRocca’s Healthspan Biology Lab and Colorado-based biotech company Sachi Bio.
“This is a novel and effective treatment to improve memory in mice,” said Prashant Nagpal, who co-founded Sachi Bio with his wife, Anushree Chatterjee. “A very important finding that we saw in this study is that you can reverse some cognitive decline. We are hoping to take this to human clinical trials next year.”
The mice behavioral tests measured memory and grip strength because grip strength and muscle function are closely linked to brain function, researchers said.
“If we can target what comes before Alzheimer’s disease, which is what this drug is meant to do, that will give people more treatment options, especially earlier in life,” Wahl said.
By next year or 2026, Nagpal hopes there will be a more conclusive data set including human trials.
“We’ve all been touched by seeing older parents and family members just being a shadow of themselves,” Nagpal said. “It’s just heartbreaking. It may seem like just a glimmer of hope, but can you latch onto it and just, you know, go for it?”
Sign up for our weekly newsletter to get health news sent straight to your inbox.
- Report an Error
- Submit a News Tip
More in Health
![By MONIKA PRONCZUK DAKAR, Senegal (AP) — The Africa Center for Disease Control and Prevention and the World Health Organization launched on Friday a continent-wide response plan to the outbreak of mpox, three weeks after WHO declared outbreaks in 12 African countries a global emergency. The estimated budget for the six-month plan is almost $600 […] By MONIKA PRONCZUK DAKAR, Senegal (AP) — The Africa Center for Disease Control and Prevention and the World Health Organization launched on Friday a continent-wide response plan to the outbreak of mpox, three weeks after WHO declared outbreaks in 12 African countries a global emergency. The estimated budget for the six-month plan is almost $600 […]](https://www.denverpost.com/wp-content/uploads/2024/09/Congo_Mpox_88825_731285.jpg?w=525)
WHO and Africa CDC launch a response plan to the mpox outbreak

Environment | Colorado gets $225,000 from CDC to measure lead, PFA exposure

Environment | Adams 14 district, parents at Dupont Elementary plan to fight gasoline storage expansion near school

Health | Coloradans using food assistance to buy produce can receive bonus matching funds
Advertisement
Supported by
In the Pacific, a ‘Dumping Ground’ for Priests Accused or Convicted of Abuse
Over a decades-long period, more than 30 Catholic priests and missionaries moved to remote island nations after they had allegedly abused children in the West, or had been found to do so.
- Share full article

By Pete McKenzie
Pete McKenzie pored over hundreds of documents, traveled across New Zealand and Fiji to report this article and spoke with survivors of clerical abuse and representatives of 20 Catholic organizations.
Pope Francis will be welcomed by children bearing flowers, a 21-gun salute and a candlelight vigil after he lands in Papua New Guinea on Friday. It will be the first papal visit in three decades to the Pacific Islands, a deeply Christian region — but one that has played a little-known role in the clergy abuse scandal that has stained the Roman Catholic Church.
Over several decades, at least 10 priests and missionaries moved to Papua New Guinea after they had allegedly sexually abused children, or had been found to do so, in the West, according to court records, government inquiries, survivor testimonies, news media reports and comments by church officials.
These men were part of a larger pattern: At least 24 other priests and missionaries left New Zealand, Australia, Britain and the United States for Pacific Island countries like Fiji, Kiribati and Samoa under similar circumstances. In at least 13 cases, their superiors knew that these men had been accused or convicted of abuse before they transferred to the Pacific, according to church records and survivor accounts, shielding them from scrutiny.
It has been widely documented that the church has protected scores of priests from the authorities by shuffling them to other places, sometimes in other countries. But what sets these cases apart is the remoteness of the islands the men ended up in, making it harder for the authorities to pursue them. The relocations also gave the men access to vulnerable communities where priests were considered beyond reproach.
Notably, at least three of these men, according to government inquiries and news media reports, went on to abuse new victims in the Pacific.
Most moved to or served in 15 countries and territories in the region in the 1990s, but one still serves as an itinerant priest in Guam, an American territory, and another has returned to New Zealand, where he has been cleared by the church to return to ministry. Both deny the allegations of abuse.
We are having trouble retrieving the article content.
Please enable JavaScript in your browser settings.
Thank you for your patience while we verify access. If you are in Reader mode please exit and log into your Times account, or subscribe for all of The Times.
Thank you for your patience while we verify access.
Already a subscriber? Log in .
Want all of The Times? Subscribe .
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
Collection 12 March 2021
2020 Top 50 Life and Biological Sciences Articles
We are pleased to share with you the 50 most downloaded Nature Communications articles* in the life and biological sciences published in 2020. (Please note we have a separate collection on the Top 50 SARS-CoV-2 papers .) Featuring authors from around the world, these papers highlight valuable research from an international community.
Browse all Top 50 subject area collections here .
* Data obtained from SN Insights (based on Digital Science's Dimensions) and has been normalised to account for articles published later in the year.

Vitamin D metabolites and the gut microbiome in older men
Here, the authors investigate associations of vitamin D metabolites with gut microbiome in a cross-sectional analysis of 567 elderly men enrolled in the Osteoporotic Fractures in Men (MrOS) Study and find larger alpha-diversity correlates with high 1,25(OH)2D and high 24,25(OH)2D and higher ratios of activation and catabolism.
- Robert L. Thomas
- Lingjing Jiang
- Deborah M. Kado

The misuse of colour in science communication
The accurate representation of data is essential in science communication, however, colour maps that visually distort data through uneven colour gradients or are unreadable to those with colour vision deficiency remain prevalent. Here, the authors present a simple guide for the scientific use of colour and highlight ways for the scientific community to identify and prevent the misuse of colour in science.
- Fabio Crameri
- Grace E. Shephard
- Philip J. Heron
Effect of gut microbiota on depressive-like behaviors in mice is mediated by the endocannabinoid system
The gut microbiota may contribute to depression, but the underlying mechanism is not well understood. Here the authors use a mouse model of stress induced depression to demonstrate that behavioural changes conferred by fecal transplant from stressed to naïve mice require the endocannabinoid system.
- Grégoire Chevalier
- Eleni Siopi
- Pierre-Marie Lledo
The default network of the human brain is associated with perceived social isolation
Here, using pattern-learning analyses of structural, functional, and diffusion brain scans in ~40,000 UK Biobank participants, the authors provide population-scale evidence that the default network is associated with perceived social isolation.
- R. Nathan Spreng
- Emile Dimas
- Danilo Bzdok
Non-invasive early detection of cancer four years before conventional diagnosis using a blood test
Patients whose disease is diagnosed in its early stages have better outcomes. In this study, the authors develop a non invasive blood test based on circulating tumor DNA methylation that can potentially detect cancer occurrence even in asymptomatic patients.
- Xingdong Chen
- Jeffrey Gole
Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure
Regulatory and coding regions of genes are shaped by evolution to control expression levels. Here, the authors use deep learning to identify rules controlling gene expression levels and suggest that all parts of the gene regulatory structure interact in this.
- Christoph S. Börlin
- Aleksej Zelezniak
A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity
Antibody mediated immunity to SARS-CoV-2 will affect future transmission and disease severity. This systematic review on antibody response to coronaviruses, including SARS-CoV-2, SARS-CoV, MERS-CoV and endemic coronaviruses provides insights into kinetics, correlates of protection, and association with disease severity.
- Angkana T. Huang
- Bernardo Garcia-Carreras
- Derek A. T. Cummings

Biomineral armor in leaf-cutter ants
Biomineral armour is known in a number of diverse creatures but has not previously been observed in insects. Here, the authors report on the discovery and characterization of high-magnesium calcite armour which overlays the exoskeletons of leaf-cutter ants.
- Chang-Yu Sun
- Cameron R. Currie
Senolytics prevent mt-DNA-induced inflammation and promote the survival of aged organs following transplantation
Organ transplantation involving aged donors is often confounded by reduced post-transplantation organ survival. By studying both human organs and mouse transplantation models, here the authors show that pretreating the donors with senolytics to reduce mitochondria DNA and pro-inflammatory dendritic cells may help promote survival of aged organs.
- Jasper Iske
- Midas Seyda
- Stefan G. Tullius
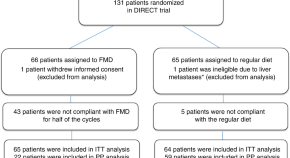
Fasting mimicking diet as an adjunct to neoadjuvant chemotherapy for breast cancer in the multicentre randomized phase 2 DIRECT trial
Preclinical evidence suggests that a fasting mimicking diet (FMD) can make cancer cells more vulnerable to chemotherapy, while protecting normal cells. In this randomized phase II clinical trial of 131 patients with HER2 negative early stage breast cancer, the authors demonstrate that FMD is safe and enhances the effects of neoadjuvant chemotherapy on radiological and pathological tumor response.
- Stefanie de Groot
- Rieneke T. Lugtenberg
- Dutch Breast Cancer Research Group (BOOG)
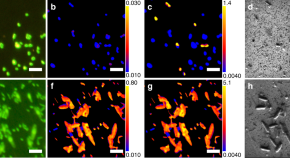
Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years
The discovery of aerobic microbial communities in nutrient-poor sediments below the seafloor begs the question of the mechanisms for their persistence. Here the authors investigate subseafloor sediment in the South Pacific Gyre abyssal plain, showing that aerobic microbial life can be revived and retain metabolic potential even from 101.5 Ma-old sediment.
- Yuki Morono
- Fumio Inagaki
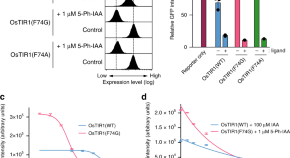
The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice
Auxin-inducible degron systems can be leaky and require high doses of auxin. Here the authors establish AID2 which uses an OsTIR1 mutant and the ligand 5-Ph-IAA to overcome these problems and establish AID-mediated target depletion in mice.
- Aisha Yesbolatova
- Yuichiro Saito
- Masato T. Kanemaki
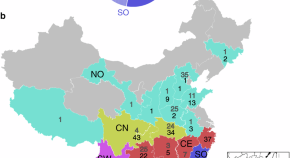
Origin and cross-species transmission of bat coronaviruses in China
Bats are a likely reservoir of zoonotic coronaviruses (CoVs). Here, analyzing bat CoV sequences in China, the authors find that alpha-CoVs have switched hosts more frequently than betaCoVs, identify a bat family and genus that are highly involved in host-switching, and define hotspots of CoV evolutionary diversity.
- Alice Latinne
- Peter Daszak

Multivariate genomic scan implicates novel loci and haem metabolism in human ageing
Ageing phenotypes are of great interest but are difficult to study genetically, partly due to the sample sizes required. Here, the authors present a multivariate framework to combine GWAS summary statistics and increase statistical power, identifying additional loci enriched for aging.
- Paul R. H. J. Timmers
- James F. Wilson
- Joris Deelen
A microsporidian impairs Plasmodium falciparum transmission in Anopheles arabiensis mosquitoes
Mircobial symbionts of mosquitoes can affect transmission of human pathogens. Here, Herren et al . identify a microsporidian symbiont in Anopheles gambiae that impairs transmission without affecting mosquito fecundity or survival.
- Jeremy K. Herren
- Lilian Mbaisi
- Steven P. Sinkins
Gene editing and elimination of latent herpes simplex virus in vivo
Herpes simplex virus establishes lifelong latency in ganglionic neurons, which are the source for recurrent infection. Here Aubert et al. report a promising antiviral therapy based on gene editing with adeno-associated virus-delivered meganucleases, which leads to a significant reduction in ganglionic HSV loads and HSV reactivation.
- Martine Aubert
- Daniel E. Strongin
- Keith R. Jerome
A predictive index for health status using species-level gut microbiome profiling
A biologically-interpretable and robust metric that provides insight into one’s health status from a gut microbiome sample is an important clinical goal in current human microbiome research. Herein, the authors introduce a species-level index that predicts the likelihood of having a disease.
- Vinod K. Gupta
- Jaeyun Sung
Bacterial nanotubes as a manifestation of cell death
Bacterial nanotubes and other similar membranous structures have been reported to function as conduits between cells to exchange DNA, proteins, and nutrients. Here the authors provide evidence that bacterial nanotubes are formed only by dead or dying cells, thus questioning their previously proposed functions.
- Jiří Pospíšil
- Dragana Vítovská
- Libor Krásný
Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment
Intermittent fasting (IF) has been shown beneficial in reducing metabolic diseases. Here, using a multi-omics approach in a T2D mouse model, the authors report that IF alters the composition of the gut microbiota and improves metabolic phenotypes that correlate with cognitive behavior.
- Zhigang Liu
- Xiaoshuang Dai
Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells
Aging involves gradual loss of tissue function, and transcription factor (TF) expression can ameliorate this in progeroid mice. Here the authors show that transient TF expression reverses age-associated epigenetic marks, inflammatory profiles and restores regenerative potential in naturally aged human cells.
- Tapash Jay Sarkar
- Marco Quarta
- Vittorio Sebastiano
Mitochondrial TCA cycle metabolites control physiology and disease
Mitochondrial metabolites contribute to more than biosynthesis, and it is clear that they influence multiple cellular functions in a variety of ways. Here, Martínez-Reyes and Chandel review key metabolites and describe their effects on processes involved in physiology and disease including chromatin dynamics, immunity, and hypoxia.
- Inmaculada Martínez-Reyes
- Navdeep S. Chandel
A deep learning model to predict RNA-Seq expression of tumours from whole slide images
RNA-sequencing of tumour tissue can provide important diagnostic and prognostic information but this is costly and not routinely performed in all clinical settings. Here, the authors show that whole slide histology slides—part of routine care—can be used to predict RNA-sequencing data and thus reduce the need for additional analyses.
- Benoît Schmauch
- Alberto Romagnoni
- Gilles Wainrib
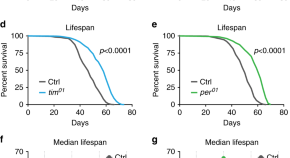
Circadian regulation of mitochondrial uncoupling and lifespan
Disruption of different components of molecular circadian clocks has varying effects on health and lifespan of model organisms. Here the authors show that loss of period extends life in drosophila melanogaster.
- Matt Ulgherait
- Mimi Shirasu-Hiza
Circadian control of brain glymphatic and lymphatic fluid flow
Glymphatic function is increased during the rest phase while more cerebrospinal fluid (CSF) drains directly to the lymphatic system during the active phase. The water channel aquaporin-4 supports these endogenous, circadian rhythms in CSF distribution.
- Lauren M. Hablitz
- Virginia Plá
- Maiken Nedergaard
Brain-inspired replay for continual learning with artificial neural networks
One challenge that faces artificial intelligence is the inability of deep neural networks to continuously learn new information without catastrophically forgetting what has been learnt before. To solve this problem, here the authors propose a replay-based algorithm for deep learning without the need to store data.
- Gido M. van de Ven
- Hava T. Siegelmann
- Andreas S. Tolias
Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy
Glioblastoma is thought to arise from neural stem cells. Here, to investigate this, the authors use single-cell RNA-sequencing to compare glioblastoma to the fetal human brain, and find a similarity between glial progenitor cells and a subpopulation of glioblastoma cells.
- Charles P. Couturier
- Shamini Ayyadhury
- Kevin Petrecca
Deep learning for genomics using Janggu
Deep learning is becoming a popular approach for understanding biological processes but can be hard to adapt to new questions. Here, the authors develop Janggu, a python library that aims to ease data acquisition and model evaluation and facilitate deep learning applications in genomics.
- Wolfgang Kopp
- Altuna Akalin

Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues
Tissue clearing has revolutionised histology, but limited penetration of antibodies and stains into thick tissue segments is still a bottleneck. Here, the authors characterise optically cleared tissue as an electrolyte gel and apply this knowledge to stain the entirety of thick tissue samples.
- Etsuo A. Susaki
- Chika Shimizu
- Hiroki R. Ueda
Sestrins are evolutionarily conserved mediators of exercise benefits
Exercise improves metabolic health and physical condition, particularly important for health in aged individuals. Here, the authors identify that Sestrins, proteins induced by exercise, are key mediators of the metabolic adaptation to exercise and increase endurance through the AKT and PGC1a axes.
- Myungjin Kim
- Alyson Sujkowski
- Jun Hee Lee
Synergistic effect of fasting-mimicking diet and vitamin C against KRAS mutated cancers
Fasting diets are emerging as an approach to delay tumor progression and improve cancer therapies. Here, the authors show that the combination of fasting-mimicking diet with vitamin C decreases tumor development and increases chemotherapy efficacy in KRAS-mutant cancer.
- Maira Di Tano
- Franca Raucci
- Valter D. Longo
Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma
Understanding the mechanisms that lead to lung adenocarcinoma metastasis is important for identifying new therapeutics. Here, the authors document the changes in the transcriptome of human lung adenocarcinoma using single-cell sequencing and link cancer cell signatures to immune cell dynamics.
- Nayoung Kim
- Hong Kwan Kim
- Hae-Ock Lee
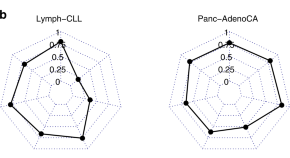
A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns
Some cancer patients first present with metastases where the location of the primary is unidentified; these are difficult to treat. In this study, using machine learning, the authors develop a method to determine the tissue of origin of a cancer based on whole sequencing data.
- Gurnit Atwal
- PCAWG Consortium
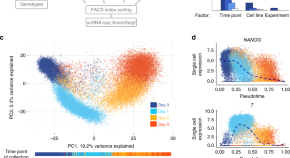
Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression
Studying the genetic effects on early stages of human development is challenging due to a scarcity of biological material. Here, the authors utilise induced pluripotent stem cells from 125 donors to track gene expression changes and expression quantitative trait loci at single cell resolution during in vitro endoderm differentiation.
- Anna S. E. Cuomo
- Daniel D. Seaton
- Oliver Stegle
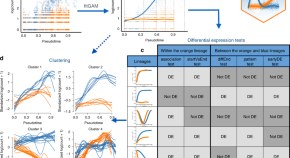
Trajectory-based differential expression analysis for single-cell sequencing data
Downstream of trajectory inference for cell lineages based on scRNA-seq data, differential expression analysis yields insight into biological processes. Here, Van den Berge et al. develop tradeSeq, a framework for the inference of within and between-lineage differential expression, based on negative binomial generalized additive models.
- Koen Van den Berge
- Hector Roux de Bézieux
- Lieven Clement

Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome
Here, Pasolli et al. perform a large-scale genome-wide comparative analysis of publicly available and newly sequenced food and human metagenomes to investigate the prevalence and diversity of lactic acid bacteria (LAB), indicating food as a major source of LAB species in the human gut.
- Edoardo Pasolli
- Francesca De Filippis
- Danilo Ercolini
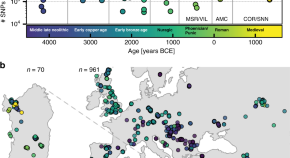
Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia
Ancient DNA analysis of early European farmers has found a high level of genetic affinity with present-day Sardinians. Here, the authors generate genome-wide capture data for 70 individuals from Sardinia spanning the Middle Neolithic to Medieval period to reveal relationships with mainland European populations shifting over time.
- Joseph H. Marcus
- Cosimo Posth
- John Novembre

Sex and APOE ε4 genotype modify the Alzheimer’s disease serum metabolome
Sex and the APOE ε4 genotype are important risk factors for late-onset Alzheimer’s disease. In the current study, the authors investigate how sex and APOE ε4 genotype modify the association between Alzheimer’s disease biomarkers and metabolites in serum.
- Matthias Arnold
- Kwangsik Nho
- Gabi Kastenmüller
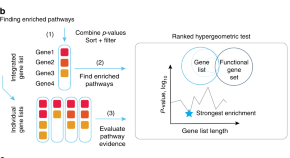
Integrative pathway enrichment analysis of multivariate omics data
Multi-omics datasets pose major challenges to data interpretation and hypothesis generation owing to their high-dimensional molecular profiles. Here, the authors develop ActivePathways method, which uses data fusion techniques for integrative pathway analysis of multi-omics data and candidate gene discovery.
- Marta Paczkowska
- Jonathan Barenboim
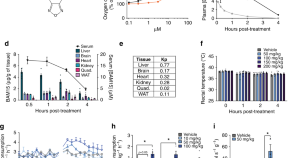
Mitochondrial uncoupler BAM15 reverses diet-induced obesity and insulin resistance in mice
Obesity is a global pandemic with limited treatment options. Here, the authors show evidence in mice that the mitochondrial uncoupler BAM15 effectively induces fat loss without affecting food intake or compromising lean body mass.
- Stephanie J. Alexopoulos
- Sing-Young Chen
- Kyle L. Hoehn
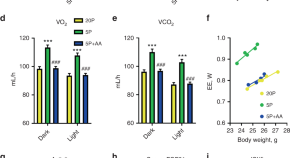
Restriction of essential amino acids dictates the systemic metabolic response to dietary protein dilution
Dietary protein dilution, where protein is reduced and replaced by other nutrient sources without caloric restriction, promotes metabolic health via the hepatokine Fgf21. Here, the authors show that essential amino acids threonine and tryptophan are necessary and sufficient to induce these effects.
- Yann W. Yap
- Patricia M. Rusu
- Adam J. Rose
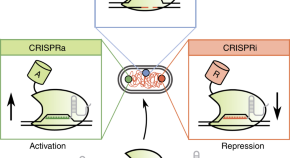
Multiplexed CRISPR technologies for gene editing and transcriptional regulation
Multiplexed CRISPR technologies have recently emerged as powerful approaches for genetic editing and transcriptional regulation. Here the authors review this emerging technology and discuss challenges and considerations for future studies.
- Nicholas S. McCarty
- Alicia E. Graham
- Rodrigo Ledesma-Amaro
Determining sequencing depth in a single-cell RNA-seq experiment
For single-cell RNA-seq experiments the sequencing budget is limited, and how it should be optimally allocated to maximize information is not clear. Here the authors develop a mathematical framework to show that, for estimating many gene properties, the optimal allocation is to sequence at the depth of one read per cell per gene.
- Martin Jinye Zhang
- Vasilis Ntranos
Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels
Ceramides are a type of sphingolipid (SL) that have been shown to play a role in several metabolic disorders. Here, the authors investigate the effect of SL-production by gut Bacteroides on host SL homeostasis and show that microbiome-derived SLs enter host circulation and alter ceramide production.
- Elizabeth L. Johnson
- Stacey L. Heaver
- Ruth E. Ley

Ancient genomes reveal social and genetic structure of Late Neolithic Switzerland
European populations underwent strong genetic changes during the Neolithic. Here, Furtwängler et al. provide ancient nuclear and mitochondrial genomic data from the region of Switzerland during the end of the Neolithic and the Early Bronze Age that reveal a complex genetic turnover during the arrival of steppe ancestry.
- Anja Furtwängler
- A. B. Rohrlach
- Johannes Krause
Brain insulin sensitivity is linked to adiposity and body fat distribution
Brain insulin action regulates eating behavior and whole-body energy fluxes, however the impact of brain insulin resistance on long-term weight and body fat composition is unknown. Here, the authors show that high brain insulin sensitivity is linked to weight loss during lifestyle intervention and associates with a favorable body fat distribution.
- Stephanie Kullmann
- Vera Valenta
- Martin Heni
Macrophages directly contribute collagen to scar formation during zebrafish heart regeneration and mouse heart repair
Macrophages mediate the fibrotic response after a heart attack by extracellular matrix turnover and cardiac fibroblasts activation. Here the authors identify an evolutionarily-conserved function of macrophages that contributes directly to the forming post-injury scar through cell-autonomous deposition of collagen.
- Filipa C. Simões
- Thomas J. Cahill
- Paul R. Riley
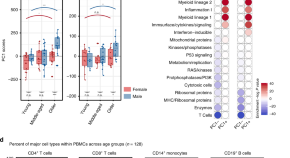
Sexual-dimorphism in human immune system aging
Whether the immune system aging differs between men and women is barely known. Here the authors characterize gene expression, chromatin state and immune subset composition in the blood of healthy humans 22 to 93 years of age, uncovering shared as well as sex-unique alterations, and create a web resource to interactively explore the data.
- Eladio J. Márquez
- Cheng-han Chung
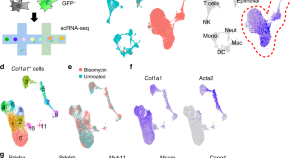
Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis
Collagen production by lung cells is critical to maintain organ architecture but can also drive pathological scarring. Here the authors perform single cell RNA sequencing of collagen-producing lung cells identifying a subset of pathologic fibroblasts characterized by Cthrc1 expression which are concentrated within fibroblastic foci in fibrotic lungs and show a pro-fibrotic phenotype.
- Tatsuya Tsukui
- Kai-Hui Sun
- Dean Sheppard
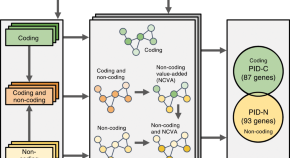
Pathway and network analysis of more than 2500 whole cancer genomes
Understanding deregulation of biological pathways in cancer can provide insight into disease etiology and potential therapies. Here, as part of the PanCancer Analysis of Whole Genomes (PCAWG) consortium, the authors present pathway and network analysis of 2583 whole cancer genomes from 27 tumour types.
- Matthew A. Reyna
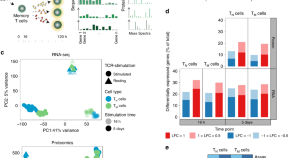
Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4 + T cells to cytokines
Cytokines critically control the differentiation and functions of activated naïve and memory T cells. Here the authors show, using multi-omics and single-cell analyses, that naïve and memory T cells exhibit distinct cytokine responses, in which an ‘effectorness gradient’ is depicted by a transcriptional continuum, which shapes the downstream genetic programs.
- Eddie Cano-Gamez
- Blagoje Soskic
- Gosia Trynka
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

COMMENTS
Diabetes mellitus (DM) is a global epidemic with increasing incidences. DM is a metabolic disease associated with chronic hyperglycemia. Aside from conventional treatments, there is no clinically approved cure... Omar I. Badr, Mohamed M. Kamal, Shohda A. El-Maraghy and Heba R. Ghaiad. Biological Research 2024 57:20.
Biological sciences encompasses all the divisions of natural sciences examining various aspects of vital processes. The concept includes anatomy, physiology, cell biology, biochemistry and ...
We hypothesized that strongly absorbing molecules can achieve optical transparency in live biological tissues. By applying the Lorentz oscillator model for the dielectric properties of tissue components and absorbing molecules, we predicted that dye molecules with sharp absorption resonances in the near-ultraviolet spectrum (300 to 400 nm) and blue region of the visible spectrum (400 to 500 nm ...
We are pleased to share with you the 50 most read Nature Communications articles* in life and biological sciences published in 2019.Featuring authors from around the world, these papers highlight ...
Biology news and videos from research institutes around the world. Updated daily. ... 2024 — New research has uncovered an extraordinary mechanism of cell division in Corynebacterium matruchotii ...
Cell biology is the discipline of biological sciences that studies the structure, physiology, growth, reproduction and death of cells. Research in cell biology uses microscopic and molecular tools ...
Read the latest Research articles from Nature Cell Biology. ... research articles. Research articles. Filter By: Article Type. All. All; Analysis (1) Article (1937) Brief Communication (210)
PLOS Biology ... PLOS Biology
Microbiomes are generally characterized by high diversity of coexisting microbial species and strains, and microbiome composition typically remains stable across a broad range of conditions. However, under fix... Sanasar G. Babajanyan, Sofya K. Garushyants, Yuri I. Wolf and Eugene V. Koonin. BMC Biology 2024 22:148.
Biology coverage from Scientific American, featuring news and articles about advances in the field. ... Replacing research animals with tools that better mimic human biology could improve medicine ...
Our biological science research journals cover a broad range of topics, from animal biology and plant science to genetic engineering, immunology, and microbiology. Read some of the latest and most talked about research from across our portfolio. Most mentioned. Read the most mentioned* articles from each of our journals in 2023 so far.
The nematode Caenorhabditis elegans is a favorite model for the study of aging. A wealth of genetic and genomic studies show that metabolic regulation is a hallmark of life-span modulation. A recent study in BMC ... Marco Gallo and Donald L Riddle. Journal of Biology 2010 9:7. Minireview Published on: 10 February 2010.
PLOS publishes a suite of peer-reviewed Open Access journals that feature quality research, expert commentary, and critical analysis across all scientific disciplines. Use these tools from PLOS to find and choose the articles you want to read: Use the search bar (above) on any journal page. Find articles by journal, subject, and other criteria.
Epigenetics research in evolutionary biology encompasses a variety of research areas, from regulation of gene expression to inheritance of environmentally mediated phenotypes. Such divergent research foci can occasionally render the umbrella term "epigenetics" ambiguous. Here I discuss several areas of contemporary epigenetics research in ...
Video: During 2023, Quanta turned a spotlight on important research progress into the nature of consciousness, the origins of our microbiomes and the timekeeping mechanisms that govern our lives and development, among many other discoveries. Revolutions in the biological sciences can take many forms. Sometimes they erupt from the use of a novel ...
Biological Research, formerly Archives of Experimental Medicine and Biology, was founded in 1964 and transferred to BioMed Central in 2014. An electronic archive of articles published between 1999 and 2013 can be found in the SciELO database.
Read the latest Research articles in Biological sciences from Scientific Reports
First published: August 26, 2024. Open Access. Wallner and Dolan describe the cell division patterns that form the flat Marchantia prothallus from a single-celled spore. A meristem with an apical stem cell develops on the prothallus to initiate indeterminate thallus growth. The transcription factor MpC3HDZ marks the first flat structure and ...
Consideration of complementary research Manuscripts that confirm, replicate, extend, or are complementary to a recently published, significant advance are still eligible for consideration in PLOS Biology and may be submitted up to six months after the first article's publication date. The complementary manuscript must present equally or more rigorous findings than the published study and ...
Current Biology is a general journal that publishes original research across all areas of biology together with an extensive and varied set of editorial sections. A primary aim of the journal is to foster communication across fields of biology, both by publishing important findings of general interest from diverse fields and through highly ...
Biology. Biology is the scientific study of life and living organisms, encompassing various sub-disciplines such as microbiology, botany, zoology, and physiology. We're dedicated to bringing you the latest research findings, innovative technologies, and thought-provoking discoveries from top scientists, research institutions, and universities ...
The Year in Biology. Momentum for new ideas in Alzheimer's research joined advances in neuroscience, developmental biology and origin-of-life studies to make 2022 a memorable year of biological insights. Our memories are the cornerstone of our identity. Their importance is a big part of what makes Alzheimer's disease and other forms of ...
The Journal of heredity. Rushworth, CA; Mitchell-Olds, T. Despite decades of research, the evolution of sex remains an enigma in evolutionary biology. Typically, research addresses the costs of sex and asexuality to characterize the circumstances favoring one reproductive mode. Surprisingly few studies address the influence of common traits ...
Shodai Komatsu. Hirohisa Ohno. Hirohide Saito. Article Open Access 17 Nov 2023 Nature Communications. Image credit: gorodenkoff / iStock. Nat Commun. Browse the 25 most downloaded Nature ...
COPENHAGEN, Denmark (AP) — Sweden's strong foraging culture could help determine how much radioactive fallout remains in the Scandinavian country 38 years after the Chernobyl nuclear explosion.. The Swedish Radiation Safety Authority has asked mushroom-pickers to send samples of this season's harvest for testing.
Research Articles are the backbone of PLOS Biology and the type of research we publish most frequently. We publish high-caliber research of any length, spanning the full breadth of the biological sciences, from molecules to ecosystems. We also consider works at the interface of other disciplines, including research of interest to the clinical ...
Divya Bhaskaran, Assistant Professor in the Exercise Science program of the Biology Department, published a research paper in the Frontiers in Neurology Journal. The article titled "Effects of an 11-week vibro-tactile stimulation treatment on voice symptoms in laryngeal dystonia" is a longitudinal clinical trial conducted during Dr Bhaskaran's post-doctoral work at the University of Minnesota.
The research came out of a partnership between CSU faculty member Tom LaRocca's Healthspan Biology Lab and Colorado-based biotech company Sachi Bio.
Michelle Mulvihill, a former nun and adviser to the Australian Catholic Church, has long accused Catholic organizations of using the Pacific Islands as a "dumping ground" for abusive priests.
Browse the 50 most downloaded Nature Communications articles across life and biological sciences published in 2020. ... these papers highlight valuable research from an international community.