- Corpus ID: 67793266

The Critical Success Factor Method: A review and practical example
- Vanessa A. Cooper
- Published 2008
- Computer Science, Business
Tables from this paper

9 Citations
Bi systems managers' perception of critical contextual success factors: a delphi study.
- Highly Influenced
Identifying Critical Success Factors through Interactive Management
Systematic literature review of critical success factors on enterprise resource planning post implementation, the other side of success factors—a systemic methodology for exploring critical success factors, critical success factors for a combined lean and iso 14001 implementation in the manufacturing industry: a systematic literature review, critical implementation factors for cloud-based enterprise resources planning in the nigerian maritime transport and supply chain, the critical success factors for electric vehicle adoption among malaysian drivers, adopting construction 4.0 to promote sustainability in the mekong delta of vietnam: a fuzzy delphi study, comunidades mediadas pela internet: fatores de sucesso e modelo de ciclo de vida, 21 references, the use of critical success factors in evaluation of information systems: a case study, sim competition paper: critical success factor analysis as a methodology for mis planning, reliability of information systems based on the critical success factors - formulation, reassessing critical success factors for erp adoption - a case study, chief executives define their own data needs..
- Highly Influential
- 11 Excerpts
Planning, Critical Success Factors, and Management's Information Requirements
Extending critical success factors methodology to facilitate broadly participative information systems planning, the alignment between organizational critical success factors and information technology capability in academic institutions, integrating management support systems into strategic information systems planning, the potentialities of focus groups in e-business research: theory validation, related papers.
Showing 1 through 3 of 0 Related Papers

- eLibrary Home
- eLibrary Login
- < Previous
Home > Affiliated Conferences > CONF-IRM > CONF-IRM 2008 > 53
CONF-IRM 2008 Proceedings
The critical success factor method: a review and practical example.
Vanessa A. Cooper , RMIT University Follow
Since the CSF method was first proposed by John Rockart in 1979, the method has been adopted for numerous research studies in Information Systems (IS). Like many research methods, the CSF method has both its supporters and critics. Almost thirty years on, this paper provides a comprehensive review of the original CSF method and of subsequent adaptations. The primary contributions and criticisms of the method are synthesized. The paper then discusses insights gained from the application of an adaptation of the CSF method in a large study involving six multi-national IT services organisations, thereby providing guidance to researchers who may consider using the method in future research.
Recommended Citation
Cooper, Vanessa A., "The Critical Success Factor Method: A review and practical example" (2008). CONF-IRM 2008 Proceedings . 53. https://aisel.aisnet.org/confirm2008/53
Since October 16, 2008
Advanced Search
- Notify me via email or RSS
- CONF-IRM 2008 Website
- All Content
Author Corner
- eLibrary FAQ
Home | About | FAQ | My Account | Accessibility Statement
Privacy Copyright

Advisory boards aren’t only for executives. Join the LogRocket Content Advisory Board today →

- Product Management
- Solve User-Reported Issues
- Find Issues Faster
- Optimize Conversion and Adoption
What are critical success factors? Examples, definition, overview

Many of the techniques, frameworks, processes, and tools in wide use today were invented during the golden era of project management in the late 1990s. With these new methodologies came a need for project stakeholders to identify key areas and actions that were required for a project to succeed.
This is where critical success factors — the key areas a product or a project need to execute or consider for a project/product to be successful — enter the picture.
What are critical success factors?
A critical success factor (CSF) is a specific element or activity that is deemed essential for an organization to achieve its mission or goal. In product management, critical success factors are the key actions a product team takes to deliver successful products that solve user problems.
Critical success factors are different from critical success criteria and key performance indicators (KPIs) . Critical success factors are action-based statements that can be assigned to an owner. Think of a CSF as a task that needs to be executed for the product to be successful.
The term critical success factors was coined in a 1961 Harvard Business Review article by Ronald Daniel titled “ Management Information Crisis .” CSFs evolved more during the late 90s to help project managers identify what needed to be done to achieve product and business goals on a bigger scale.
Who sets critical success factors?
Critical success factors are often established by product leaders, such as the VP of product or chief product officer (CPO), who own the product development process in the organization.
In product-centric organizations, CSFs are embedded into the product development process, sometimes without product managers even noticing it.
What is the outcomes hierarchy?
CSFs are the second layer of the outcomes hierarchy. The outcomes hierarchy flows as follows:
Deliverable
Critical success factors (csfs), critical success criteria (csc), key performance indicators (kpis).
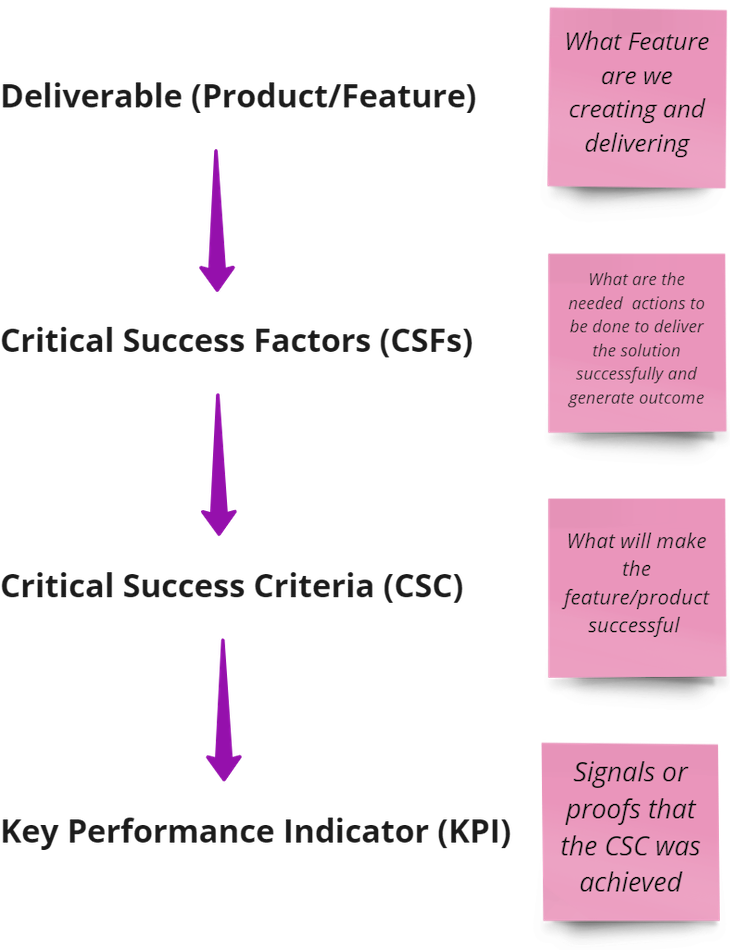
A deliverable could be any product, feature, or enhancement you are building. For a product team to build an effective feature, there should be three more layers: critical success factors (CSFs), critical success criteria (CSC), and key performance indicators (KPIs).
The critical success factor, as mentioned before, is what needs to be done to build a successful product. But it doesn’t make sense to have action steps without understanding the baseline of what success looks like. This is where CSCs and KPIs come in.
Critical success criteria are the benchmarks by which you measure the success of the feature or initiative you are pursuing. Think of it as the ultimate outcome you want to reach.
Examples of critical success criteria are increasing the number of monthly registered users, decreasing the time from searching to placing order for an ecommerce product, etc.
KPIs are a way to track your CSC quantitatively. So what signals might tell us that we achieved our CSC?
Referring back to our previous example, can we say we achieved success if only one new customer registers? Of course not.
Over 200k developers and product managers use LogRocket to create better digital experiences
Coupling your CSC with quantifiable business goals will generate a KPI — for example, increasing the number of monthly registered users by 30 percent or reducing the time from searching to placing an order by three minutes.
Benefits of using critical success factors
Setting CSFs can help you:
Promote cross-functional collaboration
Streamline product and project management processes, align stakeholders.
When the entire company — not only the product team — embraces CSFs, cross-functional teams are much better equipped to work together with minimal friction. The list of CSFs serves as a compass to help these disparate teams navigate the broader roadmap of activities required to achieve product objectives.
By embedding CSFs into the development process, the company can establish a template for success in any initiative — e.g., building features, introducing enhancements. CSFs derived from past experience will increase the likelihood of the product being successful and achieving the objective.
Having established steps to achieve success will significantly result in better stakeholder alignment. This will help the product team encounter less noise from the outside departments and will allow the outside departments and divisions to have more clarity around the holistic success process taken to maximize the feature success chances.
How to identify critical success factors
Identifying critical success factors isn’t a one-off task; it’s an ongoing initiative. While there’s no one-size-fits-all approach, the general process for establishing critical success factors is as follows:
- Start with the product strategy
- Analyze old projects
- Collaborate with product leaders
1. Start with the product strategy
Critical success factors differ from organization to organization and from product to product. Your product strategy will answer most of your questions.
The product strategy should outline your product aspirations and goals. It’s essential to keep your product strategy in mind while building your first CSFs. This will help you and your product team stay aligned.
2. Analyze old projects
Have you built a feature or product that positively impacted all your success metrics? If so, then start there.
In a workshop, identify with your stakeholders what has impacted success, what went right, and what went wrong. Also any additional insights and opinions you can collect from other stakeholders.
3. Collaborate with product leaders
Now that you have synthesized stakeholders’ insights, you can start collaborating with product leaders to develop a first draft of your critical success factors. The draft should include tailored steps or key focus areas for your product team to deliver the perfect feature or enhancement.
Start executing based on your process and your CSFs. Validate the effectiveness of those success factors and measure whether the features or enhancements delivered generate the expected results.
Based on the results, you should always tweak your CSFs to accommodate new insights and changing conditions. Treat your CSFs as an ongoing project that you will continuously refine and improve with your stakeholders and, most importantly, your product team.
Critical success factors: Examples
There are many product development process components that could serve as a critical success factor. Below are four success factors I commonly see across startups and product organizations:
Build a clear product strategy
Understand customer pain points, analyze product performance regularly, create value continuously.
A clear product strategy is the first step for any product. Your strategy will guide the product team and help them identify their customers, introduce enhancements and features, and, more importantly, prioritize them based on the strategic goals.
Building shiny new features that don’t solve customer problems is unprofitable for the business. All it does is burn resources with no real business return on investment (ROI).
However, understanding customer pain points through surveys, customer interviews, observations, contextual inquiry, and focus groups will increase your chances of delivering real value to the user and, thus, positively impacting metrics such as retention and activation.
Analyzing your performance using product analytics tools or through qualitative methods such as interviews will help you and your team create a top-notch product. Analyzing the product performance enables the product team to ease critical flows for the customer, helping them achieve their tasks and solve their problems efficiently.
Creating value in a continuous manner and delighting the user will help establish your product in the market. By solving more and more problems your users face, you increase the chances they’ll stick around.
Final thoughts
Critical success factors (CSFs) — also called key results areas (KRAs) — are crucial for any product team to achieve success. CSFs help the product team define the areas, actions, and steps that are absolutely necessary to achieve success.
CSFs also help product teams assess the areas in which they excel and areas that need improvement. By holistically detecting the faulty key success areas, they can introduce tweaks to the process and build better products during subsequent stages.
Featured image source: IconScout
LogRocket generates product insights that lead to meaningful action
Get your teams on the same page — try LogRocket today.
Share this:
- Click to share on Twitter (Opens in new window)
- Click to share on Reddit (Opens in new window)
- Click to share on LinkedIn (Opens in new window)
- Click to share on Facebook (Opens in new window)
- #product strategy
Stop guessing about your digital experience with LogRocket
Recent posts:.

Feedback management tools and strategies
Feedback management refers to a structured process for gathering and analyzing feedback to improve products, services, or processes.

An overview of feature-driven development (FDD)
FDD is an agile framework for software development that emphasizes incremental and iterative progress on product features development.

Leader Spotlight: Making the customer feel like a regular, with Judy Yao
Judy Yao talks about creating a digital experience that makes customers feel as if they were repeat, familiar customers in a physical store.

How can PMs benefit from generative AI
AI amplifies your potential when you use it as a co-pilot. However, don’t forget you’re the driver, not the passenger.

Leave a Reply Cancel reply
Understanding Critical Success Factors (CSFs) in Strategic Planning
Every business needs a roadmap for success. Without one, distinguishing victories from setbacks becomes a daunting task, casting uncertainty over the attainment of goals. Delving into the realm of Critical Success Factors (CSFs) unveils their pivotal role in steering the course of businesses and projects toward triumph.These factors serve as the guiding lights, ensuring teams and departments are synchronized, and efforts are channeled towards common objectives.
What Are Critical Success Factors?
Critical Success Factors (CSFs) are the essential elements that must be achieved to ensure success for a company or project. Understanding these factors is crucial as they help focus efforts on the most impactful areas. CSFs are not just about identifying what to do but also clarifying what not to waste resources on. They are tailored to specific industries and business models, making them unique and vital for strategic alignment.
These factors are crucial for the success of a project, initiative, or business strategy. CSFs vary depending on the industry, organization, and specific objectives, but they generally encompass the following characteristics:
Key Goals and Objectives: CSFs are directly linked to the primary goals and objectives of an organization or project. They represent the most critical aspects that must be achieved to consider the endeavor successful.
Measurability: CSFs should be measurable so that progress can be tracked effectively. They often have associated key performance indicators (KPIs) or metrics that indicate whether the factors are being met.
Strategic Alignment: CSFs align with the overall strategy and vision of the organization. They reflect the areas where the organization must excel to fulfill its strategic objectives.
Criticality : CSFs are essential for success. Failure to achieve these factors significantly increases the risk of failure for the project or organization as a whole.
Focus: CSFs help prioritize resources and efforts by highlighting the most critical areas that require attention and investment.
The Role of CSFs in Strategic Success
CSFs play a pivotal role in strategic planning by providing a clear roadmap for success. They help organizations prioritize their goals and allocate resources effectively. By defining critical success factors, companies can:
- Ensure that all team members are aligned with the strategic objectives.
- Measure progress quantitatively, as each CSF can be associated with specific performance metrics.
- Adapt to changing market dynamics by regularly reviewing and updating the CSFs.
For instance, in a technology company, a CSF might be the development of a new patentable technology, whereas, in a retail business, a CSF could be customer satisfaction ratings. This specificity ensures that strategic efforts are concentrated on the most critical areas.
Moreover, tools like Visual Strategic Planning Tools can significantly enhance the ability to visualize and manage these critical success factors, ensuring that they are not just defined but actively monitored and achieved.
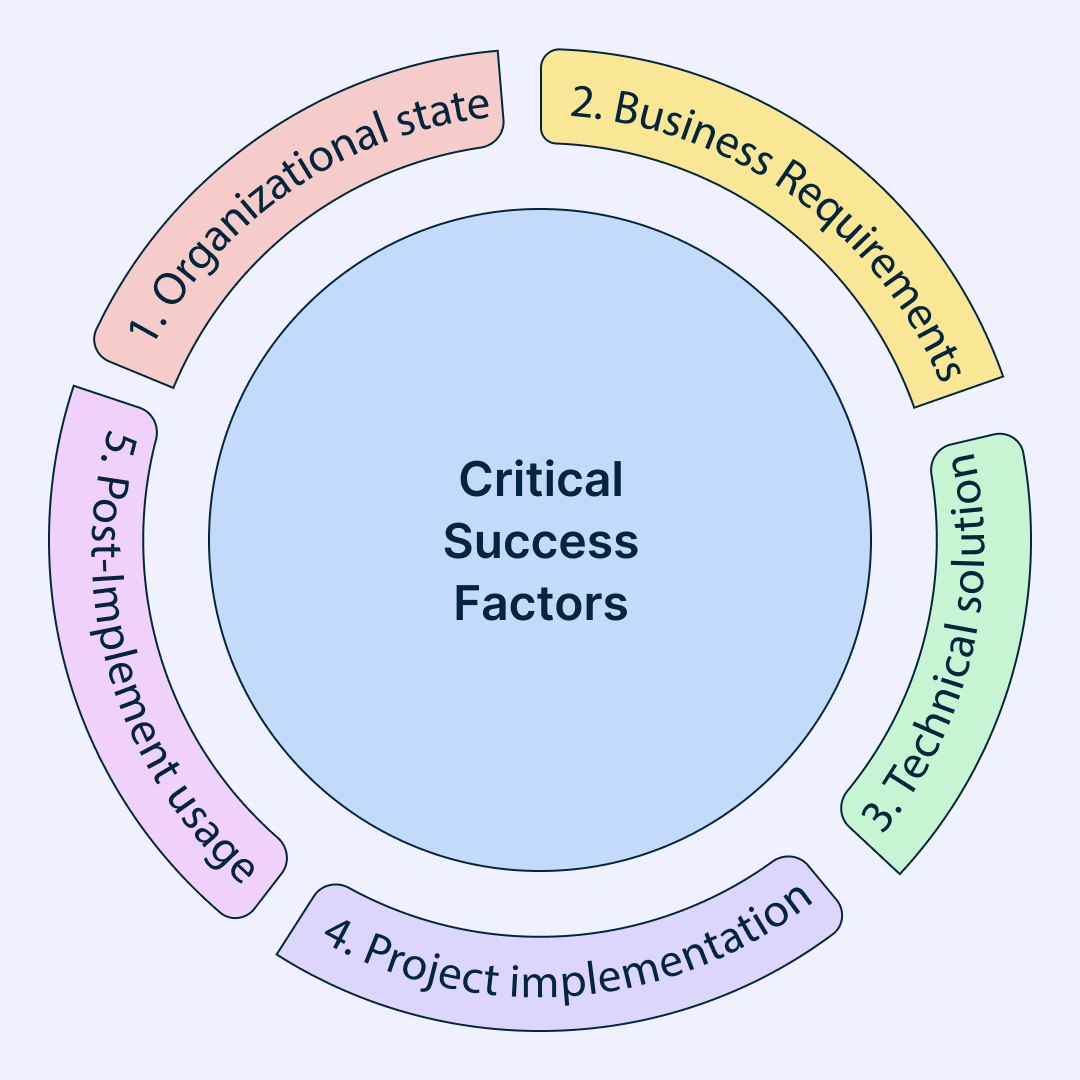
Types of Critical Success Factors:
In his seminal work, Rockart outlined four distinct categories of Critical Success Factors (CSFs), each serving as a cornerstone in the foundation of organizational triumph
Industry Factors: These stem from the unique dynamics of your industry, dictating the essential actions required to maintain competitiveness. For instance, in the realm of technology startups, innovation emerges as a pivotal CSF, driving evolution and differentiation amidst fierce competition.
Environmental Factors: Arising from broader macro-environmental forces, these factors encompass elements such as the business climate, economic fluctuations, competitor landscapes, and technological advancements. Conducting a thorough PEST Analysis unveils the intricacies of these factors, empowering organizations to navigate uncertainties with foresight and adaptability.
Strategic Factors : Tailored to the specific competitive strategy adopted by your organization, these factors delineate the strategic choices guiding positioning and marketing endeavors. Whether pursuing a strategy of high-volume, low-cost production or opting for a niche, high-value approach, strategic CSFs illuminate the pathway to sustained relevance and profitability.
Temporal Factors: Reflecting the internal flux and evolution within your organization, temporal CSFs are transient in nature, responding to short-lived barriers, challenges, and opportunities. For instance, amidst rapid expansion, a critical imperative might revolve around scaling international sales operations, highlighting the dynamic interplay between internal growth trajectories and external market demands.
Critical Success Factors (CSFs) VS Key Performance Indicators (KPIs)
Understanding the distinction between Critical Success Factors (CSFs) and Key Performance Indicators (KPIs) is crucial for effective strategic planning. While both are essential metrics in business strategy, they serve different purposes and are used in different contexts.
Critical Success Factors are the essential areas of activity that must be performed well to achieve the strategic goals of an organization. These are the elements that are critical for success in achieving the strategic objectives. On the other hand, Key Performance Indicators are quantifiable measurements that reflect the critical success factors of an organization. They are used to gauge the performance and success of an initiative, often linked directly to strategic objectives.
For instance, if a critical success factor for a tech company is ‘innovation,’ a corresponding KPI might be the number of new patents filed per year or the percentage of revenue from new products.
Using KPIs to Measure CSFs
Effectively measuring CSFs through KPIs requires a clear understanding of the strategic goals and the critical factors that drive them. Here are some ways KPIs can be used to measure the effectiveness of CSFs:
- Alignment of KPIs with strategic goals to ensure they reflect the critical success factors.
- Regular review and adjustment of KPIs to adapt to changing circumstances and ensure they remain relevant to the CSFs.
- Utilization of tools like Balanced Scorecard Templates to visualize and track these indicators effectively.
- Ready to use
- Fully customizable template
- Get Started in seconds

It’s important to avoid the pitfall of confusing CSFs with KPIs. While KPIs are indicators of performance, CSFs are the areas that determine whether the organization will achieve its strategic goals. Understanding this distinction helps organizations focus on what truly matters and allocate resources accordingly.
Building an Organizational Strategy Around Critical Success Factors
Integrating critical success factors (CSFs) into your business planning isn’t just about identifying what’s important; it’s about embedding these factors into the very fabric of your organizational strategy. This integration ensures that every decision and action aligns with your overarching goals, propelling your business towards success.
Integrating CSFs into Business Planning
Leadership plays a pivotal role in fostering a culture that prioritizes CSFs. It starts with a clear communication of what these factors are and how they tie into the daily operations and long-term objectives of the company. Here are some steps to effectively integrate CSFs into your business planning:
- Define and Align: Clearly define your CSFs and ensure they are in harmony with your organizational values and strategic goals. This alignment is crucial for maintaining focus and direction.
- Communicate: Use every opportunity to communicate the defined CSFs across all levels of the organization. This ensures everyone is on the same page and pulling in the same direction.
- Embed: Integrate CSFs into all planning documents and tools. Use frameworks like Impact Mapping Templates to visualize how individual actions and strategies connect back to these critical factors.
- Review: Regularly review and adjust CSFs to respond to changing market conditions or internal company shifts. This agility allows your business to remain relevant and competitive.
Identifying and Setting Critical Success Factors for Your Business
Identifying and setting the right critical success factors (CSFs) is pivotal for any business aiming to achieve its strategic goals. This process requires a structured approach and keen insight into both the market and internal capabilities. Here, we outline a five-step process to effectively pinpoint and refine CSFs that align with your business objectives.
- Step 1: Gather Stakeholder Input - Engage with key stakeholders from various departments to get a comprehensive view of the strategic needs and expectations. This collaborative approach ensures that the CSFs developed are inclusive and representative of the entire organization.
- Step 2: Conduct Market Analysis - Utilize resources like Scenario Planning Guide to understand market trends and competitor strategies. This analysis helps in setting CSFs that are not only relevant but also competitive.
- Step 3: Define CSFs - Based on the insights gathered, define clear and measurable CSFs. Ensure they are specific, achievable, and directly tied to strategic objectives.
- Step 4: Refine and Adjust - CSFs should not be static. Regularly review and refine them based on ongoing feedback and changing market conditions to keep them relevant and impactful.
- Step 5: Implement and Monitor - Implement the CSFs across the organization and monitor their progress. Use visual project management tools to track these factors and make adjustments as necessary.
By following these steps, businesses can ensure that their CSFs are not only defined but are also aligned with the overall strategic vision, thereby enhancing the likelihood of achieving desired outcomes. Remember, the key to successful strategic planning is not just identifying CSFs but continuously adapting them to fit the evolving business landscape.
Practical Tips for Creating Effective Critical Success Factors
Creating effective critical success factors (CSFs) is pivotal for any organization aiming to achieve its strategic objectives. Here are some practical tips to ensure your CSFs are clear, specific, and aligned with your business goals.
- Clarity and Specificity: Each CSF should be distinctly defined to avoid ambiguity. This clarity helps team members understand exactly what is expected and how it contributes to the organization’s success.
- Alignment with Strategic Objectives: CSFs should directly support the strategic goals of your organization. This alignment ensures that every effort contributes towards the overarching objectives.
Avoiding Common Pitfalls: One common mistake is setting too many CSFs, which can dilute focus and resources. Prioritize CSFs that have the most significant impact on your strategic goals.
Involving Cross-Functional Teams: CSFs should be developed with input from various departments to ensure they are comprehensive and inclusive. Engage teams through platforms that foster collaboration, such as Retrospective Meetings for Cross-Functional Teams to gather diverse insights and drive collective commitment.
Regular reviews and updates to CSFs are crucial. The business landscape is dynamic, and your CSFs should evolve to reflect changes in the market and internal business processes. Leveraging a centralized platform like Creately can facilitate the continuous monitoring and updating of CSFs, ensuring they remain relevant and impactful.
How Creately Supports Setting and Achieving Organizational Goals through CSFs
Setting and achieving organizational goals hinge significantly on identifying and leveraging critical success factors (CSFs). Creately, with its advanced visual collaboration platform, provides an array of tools designed to enhance strategic planning and execution. Here’s how Creately’s features align with the needs of organizations aiming to master their strategic objectives through effective use of CSFs.
Creately’s Tools for Strategic Planning
- Visual Canvas: Creately’s visual canvas offers a dynamic space for teams to brainstorm, map out strategies, and visualize the relationships between different CSFs. This is crucial for understanding how various factors interlink and influence overall strategic success.
- Multiple Visual Frameworks: With access to various frameworks such as Business Model Canvas Template and Strategic Planning Tools , teams can effectively define and align their organizational goals with the identified CSFs, ensuring that every action taken is strategically oriented.
Join over thousands of organizations that use Creately to brainstorm, plan, analyze, and execute their projects successfully.
More Related Articles
Chiraag George is a communication specialist here at Creately. He is a marketing junkie that is fascinated by how brands occupy consumer mind space. A lover of all things tech, he writes a lot about the intersection of technology, branding and culture at large.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- v.11(5); 2021

Original research
Success and limiting factors in health service innovation: a theory-generating mixed methods evaluation of uk projects, kathleen leedham-green.
1 Medical Education Research Unit, Faculty of Medicine, Imperial College London, London, UK
Alec Knight
2 School of Population Health and Environmental Sciences, Faculty of Life Sciences and Medicine, King’s College London, London, UK
Gabriel B Reedy
3 Centre for Education, Faculty of Life Sciences and Medicine, King’s College London, London, UK
Associated Data
bmjopen-2020-047943supp001.pdf
Data are available upon reasonable request. Due to the highly individual nature of healthcare innovations and the limited geographic area of this study, we are unable to provide our raw data. We undertake to provide a redacted data set upon reasonable request.
To explore and explain success and limiting factors in UK health service innovation.
Mixed methods evaluation of a series of health service innovations involving a survey and interviews, with theory-generating analysis.
The research explored innovations supported by one of the UK’s Academic Health Science Networks which provides small grants, awards and structural support to health service innovators including clinical academics, health and social care professionals and third-sector organisations.
Participants
All recipients of funding or support 2014–2018 were invited to participate. We analysed survey responses relating to 56 innovation projects.
Responses were used to conceptualise success along two axes: value creation for the intended beneficiaries and expansion beyond its original pilot. An analysis of variance between categories of success indicated that participation, motivation and evaluation were critical to value generation; organisational, educational and administrative support were critical to expansion; and leadership and collaborative expertise were critical to both value creation and expansion. Additional limiting factors derived from qualitative responses included difficulties navigating the boundaries and intersections between organisations, professions, sectors and cultures; a lack of support for innovation beyond the start-up phase; a lack of protected time; and staff burn-out and turnover.
Conclusions
A nested hierarchy of innovation needs has been derived via an analysis of these factors, providing targeted suggestions to enhance the success of future innovations.
Strengths and limitations of this study
- A strength of this research is that it compares many innovations in a consistent way, and that it provides insights across a range of categories of success.
- A limitation of this research is that it is situated in a single geographical context; however, repeating these methods in different contexts should produce locally relevant results.
- Few mid-level theories relating to innovation are grounded in data that include projects that have not achieved their intended outcomes; therefore, we may have identified novel insights.
- Many of the success factors we have identified are not unique to this study; however, they have been subjected to further statistical analysis and found to differentiate significantly across categories of success in this context.
- More research is needed to examine whether addressing these factors prospectively enhances the success of future innovations.
Healthcare systems worldwide are faced with increasing demand linked to the rising burden of disease within a resource-constrained environment. 1 This has led to a pressing need to find and disseminate innovative ways of meeting the healthcare needs of patients and communities in ways that are more sustainable. 2 The WHO characterises health service innovation as ‘a novel set of behaviours, routines, and ways of working that are discontinuous with previous practice, are directed at improving health outcomes, administrative efficiency, cost-effectiveness, or users’ experience and that are implemented by planned and coordinated actions’ 3 (p 7).
Academic efforts in the health sciences continue to sharpen the focus on impact, rather than the creation of ‘new knowledge’ as the primary goal of research activity. At the vanguard are implementation scientists who work to translate research and innovation into clinical practice, navigating institutional, organisational, structural and cultural complexities to improve services. 4 New support structures have emerged, such as the 15 Academic Health Science Networks set up in 2013 by the National Health Service (NHS) England, with funding streams that aim to support and encourage innovation at various levels. 5 After more than half a decade of programme grants, the impact of these innovation programmes is a legitimate subject of enquiry: how and why have certain innovations become normalised, sustained or spread, and why have others struggled or stopped?
The knowledge created through an individual innovation is likely to be complex and context dependent, providing insights that may not necessarily be generalisable. 6 Meta-analyses are faced with the complexity of synthesising multiple project evaluations that may be reported in different ways. The published literature on health service innovation contains few analyses of unsuccessful innovations, despite attempts to encourage negative reporting. 7 By evaluating a large corpus of projects across one of these academic health sciences networks in a systematic way, we have an opportunity to directly compare innovations including those that may have struggled or stopped and not reached the literature.
This study thus sets out to explore a large number of innovations, both as individual projects in their unique local contexts, and as part of a larger integrative study. By isolating the factors that differentiate between categories of success, our aim is to produce an empirically derived explanatory model, and thereby to inform and enhance the success of future innovations.
Research aim
To explore and explain success and limiting factors in health service innovation.
Methodological orientation and theory
This study is situated at the intersection of policy, social sciences and organisational research. Our philosophical assumptions are that there are real differences in the success of innovations, but also that success is fundamentally a subjective construct. Any research will only produce an approximation of the truth, and findings must be interpreted with an appreciation for context. We therefore position this research at the boundary of critical realism and constructivism. 8
Adopting Varpio et al ’s terminology on the philosophy of research, we are taking an inductive approach that works towards a theoretical framework, rather than applying a pre-existing theoretical framework to this study. 9
We have adopted what Creswell et al refer to as a sequential mixed methods design. 10 According to Creswell, insight can emerge from exploring first through qualitative methods (in our case a published qualitative review and interviews) the types of factors that might be important, and then designing questionnaires to explore their salience to a population (called an ‘exploratory sequential design’). Insight can also emerge from collecting survey data initially and then following up with interviews to help explain the survey results in more detail: an ‘explanatory sequential design’. Where both qualitative and quantitative data are collected simultaneously, one set of data can be used to triangulate the other (eg, where the meaning of one is unclear), or they can be used in complementary ways to illuminate each other (eg, one determining which factors are important, the other illuminating why that might be). Our research process involves both exploratory and explanatory aspects as well as triangulation and illumination. It is summarised in figure 1 .

Research overview. A mixed methods sequential research process to explore and explain success factors and limiting factors in health service innovation.
The Health Innovation Network (HIN) is one of the nationally funded Academic Health Science Networks set up by NHS England in 2013. It provides small grants, awards and structural support to academics, health and social care professionals and third-sector organisations, supporting service-level innovations to improve outcomes and value, including the sustainable use of resources. In addition, in the years 2014–2017, Health Education England (South London) provided investment in training and education innovation projects across healthcare settings in South London, through its strategic investment programme.
Participants and sampling
All recipients of HIN funding and Health Education England (South London) strategic investment programme funding and support during the years 2014–2018 were invited to participate. As our sample size was moderate, we aimed to analyse all responses and retrospectively determine whether the sample size was sufficient for thematic saturation and statistical inference. We achieved a priori thematic saturation for success factors (exemplar comments for each significant factor that we found) and inductive thematic saturation for limiting factors (content coded until no new themes arose). 11
Research team and ethics
The research was commissioned by HIN in collaboration with Health Education England and conducted by an independent research team at King’s College London. The research team comprised a postdoctoral educational psychologist/learning scientist (GBR), a postdoctoral occupational psychologist/health services researcher (AK) and a medical education research fellow (KLG). None were in a position of power or influence over participants, and the research was carefully designed to be conducted at arm’s length from the funding agency. Survey responses were collected anonymously and decontextualised by the research team to encourage innovators to comment critically and safely about their projects. Innovation funding was not conditional on taking part in this research. Ethical approval was granted on 26 March 2019 by the Research Ethics Committee of King’s College London (LRS-18/19–10432). Written informed consent was obtained from interviewees. Consent was implied from participation in the survey.
Patient and public involvement
No patient was involved. The primary stakeholders in this research were health service innovators who were involved in the survey design and in checking back and refining our interpretation.
Data generation methods
Survey design.
The survey design began with the extraction of potential success factors for health service innovation from a recently published qualitative systematic review. 12 This review aimed to identify all the factors and theories associated with sustainability and scale-up (capacity building) of innovations in health services research. KLG validated and expanded these factors through scoping interviews with five experienced health service innovators. The interviews started with an open exploration of what the innovator felt had impacted on the success of their project, followed by discussion on the factors identified through the literature. Personal factors were mentioned by all stakeholders in addition to the factors from the review, suggesting these may be under-reported. An additional theme (personal factors) with related subfactors was therefore included, based on these interviews. Themes and factors are listed in figure 2 . These were used to create a mixed methods nested design survey 13 using Qualtrics software (full text in online supplemental data ).

Survey scope. The survey scope was based on a qualitative review of theories and findings relating to the sustainability and scale-up of health service innovations 12 supplemented by five scoping interviews. The questions are listed in table 1 , and the full-text survey is in the online supplemental material .
Supplementary data
The survey asked respondents to:
- Categorise and describe their project’s current status (no longer running/likely to finish soon/stable at the level of the original pilot/scaled up beyond the original pilot/too early to say/other).
Analysis of variance of potential success factors across categories of success
| Factor | Average answer across all categories* | Distribution of factor is the same across categories of expansion (proven value) | Distribution of factor is the same across categories of expansion (all) | Distribution of factor is the same across categories of realised value | Interpretation and comment on secondary analysis‡ | |
| Significance† | Significance† | Significance† | ||||
| 1 | The initiative was designed to end once a set outcome had been achieved. | −0.40 | 0.274 | 0.172 | 0.317 | |
| 2 | The initiative was designed to end after defined period of time. | −0.58 | 0.011 | 0.011 | 0.288 | Unsurprisingly, even projects with high-realised value finish if they are time bound. |
| 3 | The initiative was designed to address an important healthcare need. | 1.63 | 0.765 | 0.712 | 0.135 | |
| 4 | There was public/political recognition and concern for the problem that the initiative was designed to address. | 1.23 | 0.939 | 0.66 | 0.201 | |
| 5 | The initiative was based on a strong evidence base, and it was credible that the stated benefits could be achieved through the project plan. | 1.35 | 0.696 | 0.355 | 0.299 | |
| 6 | The project was sufficiently funded. | 1.17 | 0.941 | 0.454 | 0.126 | |
| 7 | The project had sufficient infrastructure, such as buildings, office space, materials or supplies. | 1.52 | 0.952 | 0.842 | 0.613 | |
| 8 | There were sufficient members of staff with the right skills to meet the requirements of the initiative. | 1.42 | 0.003 | 0.002 | 0.013 | Skilled workforce is a critical success factor across all definitions of success. |
| 9 | Members of staff had sufficient energy and time to dedicate to the initiative. | 1.21 | 0.033 | 0.085 | 0.362 | Time and energy are critical to whether proven innovations expand. |
| 10 | There was sufficient administrative support to deliver and maintain the initiative. | 0.85 | 0.013 | 0.019 | 0.142 | Administrative support is critical to whether an innovation expands. |
| 11 | There was sufficient technical support to deliver and maintain the initiative. | 1.04 | 0.113 | 0.187 | 0.657 | |
| 12 | There was sufficient educational support to deliver and maintain the initiative. | 1.30 | 0.023 | 0.012 | 0.089 | Educational support is critical to whether an innovation expands. |
| 13 | External political or societal factors impacted negatively on the delivery of the initiative. | −0.91 | 0.191 | 0.141 | 0.005 | External political or societal factors appear critical to whether an innovation is able to realise its intended value (inconsistent exposure/response). |
| 14 | It was necessary to adapt the project so that it aligned more closely with external political or societal priorities. | −0.72 | 0.541 | 0.252 | 0.064 | |
| 15 | We had opportunities to demonstrate the benefits of this innovation within our organisation and/or to other organisations. | 1.59 | 0.237 | 0.053 | 0.02 | Unsurprisingly, innovations that were able to realise their intended value were more likely to be able to demonstrate the benefits of their innovation. |
| 16 | Steps were taken to raise the profile of the initiative, for example, through media, marketing, community engagement or publications. | 0.85 | 0.108 | 0.059 | 0.306 | |
| 17 | There are plans to replicate this innovation at other sites or spread it to other parts of the organisation. | 0.58 | 0.024 | 0.012 | 0.228 | Unsurprisingly, innovations that have become scaled up were more likely to say there were plans to spread their innovation. |
| 18 | The initiative integrated well into existing organisational structures, programmes or policies. | 1.26 | 0.012 | 0.002 | 0.059 | The ability of an innovation to integrate into existing organisational structures may be critical to whether it becomes scaled up. |
| 19 | It was necessary to adapt the initiative so that it achieved a good fit with existing organisational structures, programmes or policies. | −0.09 | 0.053 | 0.035 | 0.115 | For innovations to scale up, they may need to adapt so that they fit within existing organisational structures. |
| 20 | The host organisation was ready and able to undertake the initiative. | 1.55 | 0.262 | 0.168 | 0.721 | |
| 21 | The initiative was hampered by opposition from within the host organisation. | −1.50 | 0.037 | 0.027 | 0.398 | However valuable an innovation is, it appears unlikely to survive if it is opposed within the host organisation. |
| 22 | The host organisation lacked the necessary values/culture to support and sustain the initiative. | −1.17 | 0.265 | 0.247 | 0.888 | |
| 23 | I was released from other duties so that I could implement this initiative. | −0.43 | 0.732 | 0.893 | 0.789 | |
| 24 | I had a supportive peer network that I could discuss any issues or problems with. | 1.32 | 0.385 | 0.562 | 0.79 | |
| 25 | I was internally motivated to implement this initiative. | 1.81 | 0.425 | 0.129 | 0.034 | Innovations appear more likely to realise their value if the innovator is internally motivated. |
| 26 | I found working on the initiative personally rewarding. | 1.81 | 0.147 | 0.067 | 0.022 | Unsurprisingly, there is a correlation between an innovation realising its value, and the innovator finding it rewarding. |
| 27 | I feel I had the right skills/experience/training to implement and sustain the initiative. | 1.62 | 0.033 | 0.008 | 0.023 | The skills of the innovator appear to be a critical success factor across all definitions of success. |
| 28 | I had sufficient energy and time to dedicate to the initiative. | 1.06 | 0.209 | 0.268 | 0.498 | |
| 29 | The project had sufficient input from experts with the necessary knowledge and experience. | 1.66 | 0.134 | 0.018 | 0.021 | Expert input appears critical to both realisation of value, and to whether it expands. |
| 30 | The outcomes and impact of the project were measured or assessed. | 1.37 | 0.060 | 0.125 | 0.119 | |
| 31 | We were able to demonstrate the effectiveness of the project. | 1.38 | 0.463 | 0.185 | 0.015 | Unsurprisingly, innovations that were able to realise their intended value were more likely to be able to demonstrate the effectiveness of their innovation. |
| 32 | Performance data were gathered and reported on a regular basis. | 1.04 | 0.346 | 0.266 | 0.159 | |
| 33 | Steps were taken to systematically improve and adapt the project. | 1.44 | 0.293 | 0.153 | 0.377 | |
| 34 | There was ongoing orientation and training available, for example, to new staff or to build capacity. | 0.87 | 0.034 | 0.03 | 0.153 | The availability of ongoing training may be critical to whether successful innovations scale up. |
| 35 | Staff were given time/incentives to attend the necessary training. | 0.47 | 0.096 | 0.159 | 0.178 | |
| 36 | Staff were required to attend the necessary training | 0.11 | 0.348 | 0.271 | 0.767 | |
| 37 | The initiative was difficult or complex to deliver. | −0.09 | 0.294 | 0.163 | 0.158 | |
| 38 | The initiative helped to make things easier or more efficient. | 0.76 | 0.86 | 0.953 | 0.182 | |
| 39 | The initiative did not require special or extra effort. | −1.09 | 0.979 | 0.869 | 0.597 | |
| 40 | I believe that the staff delivering the initiative found the work/tasks rewarding and satisfying. | 1.59 | 0.368 | 0.456 | 0.743 | |
| 41 | The project team worked well together. | 1.74 | 0.416 | 0.796 | 0.893 | |
| 42 | There were clear responsibilities for individuals the work was shared across the team. | 1.45 | 0.945 | 0.533 | 0.066 | |
| 43 | Project was overly dependent on a particular individual or individuals. | 0.57 | 0.708 | 0.355 | 0.29 | |
| 44 | I believe that the team understood what the project was trying to achieve and that it would lead to improved processes and outcomes. | 1.62 | 0.218 | 0.165 | 0.772 | |
| 45 | There were rewards or incentives that supported engagement with, and continued delivery of, the initiative. | 0.07 | 0.45 | 0.638 | 0.228 | |
| 46 | The activities and roles of the initiative were incorporated into job descriptions. | −0.30 | 0.29 | 0.243 | 0.141 | |
| 47 | Staff had time within their working hours to complete the tasks of the initiative. | 0.59 | 0.251 | 0.138 | 0.328 | |
| 48 | The initiative had leadership and/or champions who were committed and capable. | 1.62 | 0.003 | 0.001 | 0.006 | Leadership appears to be a highly significant success factor across all definitions of success. |
| 49 | There was an appropriate balance of power between those involved with the initiative. | 1.15 | 0.697 | 0.775 | 0.929 | |
| 50 | Team members were able to express their opinions, and their opinions were valued. | 1.91 | 1 (no variance) | 0.049 | 0.026 | Distributed decision-making may be a critical success factor across all definitions of success. It was common to all innovations of value that scaled up (hence no variance). |
| 51 | There was a sense of ownership and commitment by those involved with the initiative | 1.79 | 0.284 | 0.177 | 0.102 | |
| 52 | Staff who were responsible for delivering the initiative were involved as partners, and were able to shape the initiative. | 1.74 | 0.306 | 0.176 | 0.031 | Participatory processes with staff may be critical to the ability of a project to realise its intended value. |
| 53 | The beneficiaries (patients/service users) were involved as partners, and were able to shape the initiative. | 0.83 | 0.45 | 0.139 | 0.027 | Participatory processes with patients/service users may be critical to the ability of a project to realise its intended value. |
| 54 | The community in which it was situated was involved as partners, and was able to shape the initiative. | 0.96 | 0.177 | 0.034 | 0.023 | Participatory processes on a community level may be critical to both the ability of a project to realise its value and its scalability. |
| 55 | There was a collaborative network of people/organisations that helped to support and sustain the initiative. | 1.30 | 0.008 | 0.007 | 0.003 | The support of a collaborative network of people/organisation may be highly significant to both value creation and scalability. |
| 56 | It felt as though the initiative was imposed on us and there was little sense of ownership or commitment to the project. | −1.64 | 0.684 | 0.488 | 0.326 |
Analysis of variance of potential success factors across categories of success.
*Respondents on average ↑ =agree, ↗ =somewhat agree, → =neither agree nor disagree, ↘ =somewhat disagree, ↓ =disagree.
†Asymptotic significances are displayed. The significance level is 0.05. The darker the shading, the safer it is to reject the null hypothesis. Significance <0.05 indicates >95% certainty that the difference between categories is not random.
‡Secondary analysis examined the direction of the association and the strength of effect across categories of success.
- Describe the status of their project and provide qualitative insights into each of the nine themes.
Our five stakeholders helped to improve the clarity, acceptability and usability of the survey questions and instructions.
Survey distribution
A neutral administrator from Health Education England distributed the survey by email in August and September 2019 to all 176 named recipients of HIN and Health Education England (South London) funding awards, grants and bursaries. A reminder was distributed 4 weeks later to participants who had not responded. Projects that had received more than one award were sent a single survey, and participants who had run more than one project were sent a separate survey for each project.
Stakeholder follow-up interviews
KLG checked back our results and interpretation with five stakeholders identified by HIN as experienced innovators, one of whom was also involved in the original scoping interviews. Interviews lasted 30–45 min and transcription was facilitated by automated software (otter.ai). These stakeholders helped to refine the model and confirmed its applicability and utility in their context. No new themes arose; however, quotes were used to enrich our survey data.
Data analysis methods
Development of categories of success.
KLG and AK categorised projects into grades of success based on how the respondent self-categorised their project, triangulated against their qualitative survey responses. The categories of success were derived through an iterative process, involving both researchers agreeing a descriptive summary of the status of each project (eg, scaled down despite achieving better than expected patient outcomes; scaled down because the intervention did not achieve its aims). We grouped projects with similar project outcomes together, and through a process of constant comparison 14 constructed a categorisation framework that accounted for all the cases in the set.
Determination of salience of success factors
We adopted an exploratory approach to data analysis, which aims to generate rather than test theory. 15 KLG conducted an analysis of variance (ANOVA) for each of the scored factors (Kruskal-Wallis non-parametric ANOVA on rank using IBM SPSS V.25) to see whether there were significant differences between categories of success. The Kruskal-Wallis test does not assume a normal distribution in the data and can be used when the data are ordinal, for example, Likert scores. For asymmetric group sizes, the non-parametric Kruskal-Wallis test performs better than the parametric equivalent ANOVA method. 16
For each factor that was identified as being significantly different between categories of success, we conducted a secondary analysis (box plot for each category) to confirm the direction and consistency of the association. This is generated automatically by SPSS after a Kruskal-Wallis test. A graded ‘exposure-response’ relationship across all grades of success would be expected if a factor genuinely drives success. 17 Where a graded relationship was not present, this is discussed in table 1 .
Illumination of success factors
KLG and AK extracted quotes from the survey and interviews relating to each significant success factor to generate a rich description within each theme.
Inductive analysis of limiting factors
KLG coded the content of all qualitative data relating to challenges within projects that had not achieved their intended outcomes or that had scaled down or stopped (n=21) facilitated by NVivo V.12 software. GBR and KLG refined the codes and both authors worked together to inductively arrange the content into themes. 18
Development of final model
We mapped significant factors onto a 2×2 grid using a natural logarithmic scatter plot so that factors that were significant to one dimension of success were mapped to the right half of the grid, factors that were significant to a second dimension of success were mapped to the top half and factors that were significant to both were mapped to the top right quadrant. We grouped success factors into themes through a process of collaborative discussion, and we explored which themes predominated in each quadrant to generate our model which was checked back with stakeholders.
Descriptive summary
We received 63 responses, but seven were incomplete or duplicate so a total of 56 responses (31.8% of 176) were included in the analysis. Each response related to a different innovation project. Survey respondents self-identified within one or more of the following groups: the project leadership team (n=54); service delivery team (n=9); training team (n=9); administrative team (n=6); service lead (n=2); and patient/service user (n=1). Several respondents identified within multiple groups.
Projects were situated in secondary care (n=19); community care (n=14); academic sector (n=5); mental health sector (n=4); online (n=4); primary care (n=3); and the hospice sector (n=2), with the remainder working at the interfaces between services, or across sectors. Their scope ranged from national programmes at hundreds of sites, local programmes supporting tens of thousands of patients, to small intensive innovations working in new ways with a few dozen complex patients, and their duration ranged from 1 to 5 years. The innovation areas related to new ways of working in end-of-life care; disability enablement; support for complex or vulnerable patients; discharge support; pain management; patient safety innovations; recovery and rehabilitation; personalised care; chronic conditions; new models of integrated health and social care; health promotion; and novel simulation and workforce development strategies. Typical projects can be explored at the HIN website 19 ; however, for reasons of confidentiality, we cannot specify which were included in this study.
Categories of success
Our emergent framework categorised each project’s success across two dimensions: the first relating to whether the innovation was reported as generating more or less than its anticipated value for patients/carers (‘value creation axis’), the second according to whether the project became sustained or scaled up beyond the initial pilot, or whether it was scaled down or stopped (‘expansion axis’). Innovations that were within the scope and intentions of the original pilot were positioned centrally. We initially scored projects into five categorisations across the expansion axis, as some projects expanded locally and some nationally; however, there were not enough projects in each group and statistics became unreliable, so we made a pragmatic decision to adopt relative rather than absolute categories.
The resulting categorical framework is illustrated in figure 3 , with the number of innovations in each category shown in brackets.

Framework for categorisation of success within health service innovation Health service innovations were categorised across two dimensions of success through an inductive classification process. The horizontal axis relates to how successful innovations were in realising their intended value: ‘did it create more or less than its intended value for beneficiaries?’ The vertical axis relates to expansion: ‘was the innovation scaled up or scaled down from the original pilot?’ Numbers in brackets indicate how many innovations were found in each category.
Success factors
Our analysis compared the variance of success factors across innovations that had demonstrated lower than expected value (n=11), value as expected (n=25) and higher than expected value (n=20). Next, we compared variance across innovations that had diminished in scope or stopped (n=17), innovations that were running as expected (n=16) and innovations that had scaled up (n=23). Finally, we excluded low-value projects and analysed again across the expansion axis (n=10, n=12 and n=23, respectively), seeking to explore why innovations with proven value had not been scaled up.
Our analysis is presented in table 1 with significant results (p<0.05) shaded in green. At this level of significance, there is a 1 in 20 probability that a result is in fact random. We have used lighter shading to indicate factors that might potentially be significant, or which could be found not to be significant if the power of the study was increased. The final column gives our interpretation of the more significant findings (p<0.05) that takes into consideration our secondary analysis.
Many factors were similarly scored across all categories of success, for example, information technology (IT) infrastructure. This does not mean that these factors are not important, only that they were experienced similarly across all categories of success and are therefore unlikely to be the underlying cause of the relative success or failure of a project.
We have collated the significant factors together in table 2 with illuminative quotes, and we discuss both positive and negative findings within each of the nine survey themes below.
Factors that are significantly associated with innovation success with illustrative quotes
| Significant factors by theme (significant to value or expansion) | Illustrative quotes |
| Theme 1: Project aims | None of the factors related to the aims of the project were significant. |
| Theme 2: Resources and support | ‘The programme support sat with one individual rather than a team and as highlighted previously the administrative/programme support hadn't been entirely appreciated/factored in at the outset of the programme.’ (R16) ‘There are innovators out there who are doing things on their own, and the person I'm thinking about is not in a very good place. He’s got virtually no support, and I don't know how he does it.’ (FI2) ‘The resources needed in terms of administration and support were underestimated. We input far more time and admin resources than originally planned.’ (R15) ‘The envelope of funding available did not enable us to fully develop a training package which was what we had initially hoped to do.’ (R25) |
| Theme 3: How the project interfaced externally | ‘The project piggy-backed on the current social movement highlighting the needs of mental health.’ (R18) ‘Hearing has always been the poor relation to other health issues even though everyone knows someone struggles with their hearing either family or friends.’ (R35) |
| Theme 4: Organisational factors | ‘The project was presented in [area] Council, [area] NHS Trust, to the public health team in the council and the voluntary sector in [area]. It was aligned with local priorities and local initiatives. A journal article is being drafted.’ (R13) ‘There was also a disconnect between commissioner priorities & [ ] priorities in relation to the programme, which impacted on its sustainability & roll-out.’ (R16) |
| Theme 5: Personal factors | ‘I was very motivated to implement this project which was demanding as I had no protected time for it. Nonetheless, you do what is needed to achieve a goal.’ (R18) ‘I am very proud of our achievements and that the work has become a routine part of our culture and system of working.’ (R28) ‘On reflection I needed to lead this project more strongly. I tried to be facilitative and not prescriptive, but the staff were not experienced enough to utilise this opportunity. They needed more direction and support. There was some conflict between the educators and the substantive staff.’ (R10) |
| Theme 6: Project management | ‘As the project was run within [ ] and [ ], there was plenty of expertise to call upon as required.’ (R6) ‘The ongoing reporting allowed for the results to be understood early in the project, shared and used in the strategy for delivering education across [ ].’ (R5) ‘A robust evaluation was undertaken of the programme, along with regular review points to inform adaptations/opportunities for improvement.’ (R16) |
| Theme 7: Tasks of the project | No factors relating to the tasks of the project were significant. |
| Theme 8: Team processes | ‘We always express the value of our service users, administrator, and other members of the team and meet regularly to discuss well-being, progress, and evaluation.’ (R33) ‘The team got on. It was a lovely team and dynamics. We all believed in the idea and were excited about it. Obviously the project needs expertise in [ ], so in that regard the work was dependent on the availability of such expertise within the team.’ ‘Leadership hasn't been invested in providing the platform for the workshops & curriculum to be rolled out. Lip service given by leadership.’ (R56) ‘This programme was carried out by a team but lead by myself. Other parties lacked the time and incentive to commit to running and leading the programme after the 12 months of my time being project lead.’ (R46) |
| Theme 9: Collaborative and participatory practices | ‘We've had some sort of service user involvement all the way through… that’s really improved the way we’ve reflected and talked… it gives a genuineness to what we're trying to achieve… the fact that you go and work with the associates and carers, you actually go and look at the environment, you spend time with the nurses that you’re going to be teaching and all the other people that you’re working with, really helps to, you know, it definitely adds value to the project.’ (FI3) ‘There was a single practitioner using the resource on her own, and now it is nationally recognised… Without that level of support: the time, the people and the resources, we wouldn’t have got anywhere. It’s been a fantastic level of support. They designed an implementation toolkit to support practitioners embedding the programme locally.’ (FI2) ‘The team at the [ ] were fabulous and so supportive.’ (R12) |
The nine themes and factors were derived from a qualitative review of the literature supplemented by stakeholder interviews. Significant factors were derived through a survey that explored salience of factors to outcomes. Survey respondents are indicated by R(n) and follow-up interview respondents by FI(n).
Project-related factors
Interestingly, the aims of the project did not appear to be critical to success. Both successful and unsuccessful innovations were similarly reported as being designed to address an important healthcare need that was concerning to the public. All funded projects were required to articulate a credible evidence base arguing that stated benefits could be achieved through the project plan.
Resourcing and expertise
All significant resource-related success factors were associated with the workforce. Non-critical factors included infrastructure (such as buildings, materials and supplies), which were reported as sufficient; IT, which was moderately good across all categories of success; and funding issues, which were also similar across all categories of success. However, having the right numbers of staff with the right skills appeared to be highly significant, both in terms of the project being able to realise its intended value, as well as for it to become scaled up beyond the original pilot. Staff with time and energy appeared critical to whether successful innovations became scaled up, as were administrative and educational support, including the availability of ongoing educational support (eg, orientation and training for new staff, or to build capacity). Expertise appeared to be critical across all categories of success, both in terms of the innovator feeling they had the right skills, experience or training; the project having access to staff with the right skills; and having external expert input where needed.
External factors
Alignment to societal needs appeared to correlate with whether a project was able to realise its intended value, but less so with its expansion. However, as the effect size did not grow consistently across categories of value creation, we cannot necessarily infer a causal relationship. 17 Qualitative comments indicated that projects that were able to align themselves to current political or societal agendas, such as mental health, were more successful. Conversely, those attempting to work in relatively less topical areas of practice described difficulty securing strategic funding, so we have tentatively included this factor in our model.

Organisational factors
Our analysis of organisational factors indicated that the ability of an innovation to integrate into existing organisational structures, programmes or policies may be critical to whether it scales up, and possibly also to its ability to create value (p=0.059). Successful projects described adapting where necessary to achieve a good fit within organisational priorities. For the most part, host organisations were described as having a positive learning culture and were ready and able to undertake innovative initiatives; however, even innovations with proven value were unable to survive if there was opposition within the host organisation.
Personal factors
Few respondents reported being released from other duties so that they could implement their initiative. However, most respondents said they benefited from a supportive peer culture. Respondents who were able to realise value were significantly more like to say they were internally motivated and found working on the project rewarding.
Project management
Most projects measured or assessed the outcomes and impacts of the project, though this appeared to be more common in successful projects (p=0.060). Projects with high value were able to demonstrate and share this success. Leadership appeared to be a highly significant success factor across all categories of success, with struggling or unsuccessful projects citing leadership failures.
The tasks of the project
Similar to theme 2, which explored the aims of the project, the tasks of the project did not appear to be significantly different across categories of success.
Collaborative and participatory practices
Valuing team members’ opinions was highly significant across all categories of success and was present in all projects that were scaled up (hence variance not calculable). Participatory approaches were significantly associated with the ability of an innovation to generate value. These participatory processes related to the staff delivering the innovation, the intended beneficiaries and the communities in which the innovations were situated.
Finally, one of the most significant differentiating factors across all categories of success was engagement with a collaborative network that helped to support and sustain the initiative.
Limiting factor analysis
In addition to the above success factors, which were quantitatively identified, the following limiting factors were identified through our qualitative analysis of failed or struggling projects. As our limiting factor analysis is qualitative and interpretive, we present our data in line with our analysis.
Boundaries between commercial, voluntary and public sectors
While UK healthcare is primarily publicly funded and provided by the NHS, social care is often commercially provided, 20 creating the potential for friction at the interfaces between these sectors.
As the care homes are private businesses, there was some lack of political will to embrace the training, as there was a view that although there was the potential to improve health outcomes for the residents, the manager did not feel there were sufficient resources to implement the required training. (R4)
Commercial organisations were reported as unwilling to release staff for training unless the value of that training was felt within the organisation. Valuable initiatives by the voluntary sector to train social care staff, but which provided benefits in the healthcare sector, fell between sectors and were potentially unviable without direct funding.
The voluntary sector is happy to participate but there is no spare capacity within it unless there is a financial package that can go with it. (R35)
There was concern that privately funded organisations were not subject to the same standards and mandates as the publicly funded bodies, and were failing to invest in training.
Because it is not mandated, organisations do not have to engage with or release staff for education. (R2)
Restructuring within the NHS has created a set of semiautonomous institutions and organisations with different and sometimes competing priorities. 21 Some participants described difficulty aligning project aims to multiple organisational goals.
There were tensions between the two boroughs in relation to approach & resourcing. There was also a tension between commissioner expectations and practice/federation expectations which have impacted on the programmes sustainability. (R16) So, this intervention has a good return on investment, for every £1 you spend you get a return of £5.20. And they’ll say, I’m the one making the investment, but he’s the one making the return here. I’ve got a budget; he’s got another budget. We might both be in the health system, but I’m not going to spend my money if he’s the one getting the return. (FI2)
Workplace cultures and priorities
Some projects reported finding non-healthcare workers receptive to health-related training; however, some failed or struggling projects found this a challenge.
Medicines delivery teams unwilling to take on additional role. (R61) There were concerns raised by care home managers that the initiative would cause undue responsibility on individuals to make clinical decisions. (R16)
Participants described differences between academic and workplace learning cultures, and variable receptiveness of front-line clinical staff to change. Some described resistance to outsiders telling healthcare workers how to improve. This may reflect the inverse of high-value projects, which were found to engage in participatory practices, engaging patients, front-line staff and communities in codesigning their innovation.
It has been difficult to embed these products due to structural issues within the staff teams. (Nursing) It was clearly not seen as a priority. (R20) I think the main insight I would have is that when working with mental health nursing teams the researcher and research team needs to be fully integrated into team(s) and seen as part of the culture. Being an outsider does not seem to work as day-to-day practice seems to regulate research. (R20)
Participants also described tensions between management priorities and the priorities of those working directly with patients.
No interest on part of management. I don’t think they have even read it. (R57) The initiative was welcomed at service level, however there was little interest at senior management level. (R52) There is such a dislocation between commissioning and what is happening on the ground. (FI2)
Lack of support beyond the start-up phase
Participants noted ongoing privileging of new innovation over sustaining or scaling up innovations that have already demonstrated their value. For example, clinical academics do not gain publications for ongoing maintenance of innovative practice:
‘Research remit probably wouldn’t cover [further dissemination] unless there was a good likelihood of REFability’ [‘REF’ refers to the Research Excellence Framework, a scoring system used to fund the university sector]. (R1)
Participants described innovation funding streams, but articulated difficulty securing funding beyond the start-up phase.
The project was resourced sufficiently for the pilot. However, once the pilot finished so did the project. (R5) The education faculty and funding is driven towards innovation and not sustainability—this de-incentives individuals from continuing with existing projects. (R8)
Burn-out, turnover and lack of protected time
Participants described projects that were limited by staff burn-out, turnover and a lack of protected time.
My commitment to the project was there however the resources I had to continue with project were limited due to competing pressures on my time. (R12) The programme required more administrative support than anticipated & this ended up being an ask over & above someone’s day job for a prolonged period of time. (R16) The most important person was our pharmacist who moved from the pharmacy a few months after we started! (R61)
Risk as integral to innovation
Finally, it is worth noting that participants felt that risk was a necessary ingredient of healthcare innovation. Innovations that fail to demonstrate value should be supported in folding without hesitation, and lessons shared.
The project demonstrated that this initiative was not a model that would work in the hospital environment hence could not be embedded. (R30)
Final framework
We created two 2×2 matrices containing all the significant success factors across each dimension of success, shown in figure 4 . The matrix on the right excluded low-value projects in the calculation of factors significant to expansion and served to support the inclusion of some marginal factors in the final model as they became more significant despite lower power.

Critical success factors plotted according to their salience to success. Critical success factors plotted according to their significance to success on a natural logarithmic scale so that factors above and to the right are probably significant (p≤0.05). The expansion axis indicates significance to whether a project is scaled up or down beyond the original pilot. The value creation axis indicates significance as to whether it creates more or less than its intended value for beneficiaries.
Figure 4 shows clearly congruent clusters of factors in each quadrant, indicating that some types of factors may be more important to expansion, while others are more important to value creation. These clusters relate to skills and expertise, leadership and motivation, organisational fit and structural support, societal alignment and participation, and evaluation.
As outlined in table 1 , there are questions as to whether evaluation and motivation are dependent rather than independent variables: does finding working on a project personally rewarding drive success or vice versa, and does a positive evaluation drive success or vice versa? Triangulation with qualitative comments (in table 2 ) suggests that evaluation and motivation may drive success, so they have been tentatively included in our final model.
Themes that were predominantly related to value creation (participation, motivation and evaluation) were labelled value creation factors. Themes that were predominantly significant to expansion (organisational fit and structural support) were labelled expansion factors. Themes that were significant to both axes (expertise, leadership and a supportive network) were labelled core success factors. We arranged success factors into a nested hierarchy, as innovations that do not generate value are unlikely to be scaled up. Our final model in figure 5 also lists potential limiting factors identified through our inductive qualitative analysis.

Nested hierarchy of success factors and limiting factors for health service innovation. Factors that may be significant to both value generation for the intended beneficiaries and to whether the innovation is scaled up beyond the original pilot are labelled as core needs. Factors that may be significant specifically to value generation are the next priority (middle layer) as innovations that do not generate value will not become embedded or spread. Finally, factors that may primarily be significant to expansion are presented in the outermost layer. Additional limiting factors were identified through an inductive analysis of qualitative responses from projects that had scaled down or failed to produce their intended value.
This analysis of 56 health service innovation projects has enabled us to propose a model for understanding success in health service innovation that has two discrete axes: one relating to whether or not the innovation created value for its intended beneficiaries; the other relating to whether or not it was scaled up beyond the original pilot. Comparing projects across these dimensions of success has enabled us to hypothesise that:
- The core drivers of success are leadership and collaborative expertise (leadership skills and commitment, expert input, sufficient staff with the right skills and expertise, and a supportive collaborative network).
- The drivers of value creation for the intended beneficiaries are participation, motivation and evaluation (involvement of patients, public, practitioners and communities, alignment to societal needs, internal motivation, finding the project work rewarding, ability to demonstrate benefits and having opportunities to share impacts).
- The drivers of sustainability and scale-up are organisation fit and structural support (organisational fit and alignment, administrative and educational support, staff with time and energy).
Additional limiting factors included difficulties at the boundaries and intersections between organisations, professions, sectors and cultures; a lack of structural support beyond the start-up phase; and staff burn-out and turnover.
Within healthcare services, the issue of diffusion and sustainability of innovation has received widespread academic attention pioneered by Greenhalgh et al 22 who drew on Rogers’ seminal text on diffusion of innovations. 23 There have been many subsequent notable academic contributions. 24–28 Nilsen proposed an overarching framework of healthcare implementation theories according to the aim of the theory. 29 Theories such as those about innovation sustainability, which include the diffusion of innovation theory, were categorised as ‘determinant frameworks’, as they posit general types of factors that can influence the success of an innovation. We believe that our findings contribute through empirical evidence to theoretical development at this level, and thus may have wider implications than programme-level data would normally allow. According to Nilsen, such theories have typically been analysed and formulated across individual studies, at the level of meta-analysis or review 29 and may therefore be one or more steps removed from the underlying data. This study is different in that we have developed mid-range theory that is empirically grounded in programme-level data, and there is a clear line between our data and the generated theory.
Conceptions of innovation success tend to focus on sustainability 30 and scale-up. 24 We suggest that both are contingent on the ability of an innovation to provide value to its intended beneficiaries in the first place. There are few theories grounded in empirical data that explain this dimension of success. Our findings highlight the importance of patient, public and practitioner involvement, alongside the core success factors of leadership and collaborative expertise. We suggest that these are fundamental preceding factors to either sustainability or scale-up.
This ‘value for intended beneficiaries’ dimension of success also allows us to conceptualise a valuable innovation that is not growing or expanding. This, we argue, is important: healthcare innovations may have parameters within which growth and expansion are constrained, perhaps because their aims have been achieved, or because the context changes. An innovation that has met its aims but has not expanded beyond its natural boundary should be properly positioned as such.
Our ability to research a set of potentially valuable projects that were scaled down or stopped, many of which never reach the literature, may have afforded novel insights. Fixsen et al suggest that sustainability can only be asserted when the funding to support implementation is withdrawn. 31 Wiltsey Stirman et al ’s systematic review suggests sustainability can be asserted after a period of 2 years. 32 Our findings suggest that continued structural support, particularly organisational, administrative and educational support, may be critical to a project’s sustainability and scalability, and that their withdrawal may destroy potentially valuable innovations.
Finally, our findings further validate the work of Dopson et al , 33 whose qualitative exploration of a similar set of health service innovations highlighted the importance of context and process over content: it is not so much what you are trying to achieve, it is how you do it and the organisational and interpersonal contexts that you work within that matter.
A limitation of this research is its highly contextual nature. Our results may not be generalisable to all contexts; however, repeating these methods may produce locally relevant results. The ANOVA depends on the universe of potential factors having been correctly identified and a large enough number of innovations to produce statistical significance. The research could be improved by more extensive validation of factors, patient and public involvement, further testing the directionality of tentative factors, a greater geographical spread and a greater number of projects to allow for finer grading across the expansion axis.
Our findings suggest that organisations and policy makers wishing to support service-level innovation in similar healthcare contexts address the factors identified through this research as critical to success.
Such strategies might include:
- Supporting innovators with the right skills and expertise, including leadership skills, implementation support and evaluation expertise.
- Innovation networks to provide opportunities to showcase success and provide a peer community of expertise and support.
- Emphasising participatory practices and collaborative approaches, so that innovations are more likely to align to societal and organisation goals and generate value for patients, communities and practitioners.
- Providing administrative and educational support during the scale-up phase, and ensuring that this support is maintained or handed over rather than withdrawn to schedule.
- Recognising and enhancing the internal motivation and drive of innovators as well as more goal-oriented motivations such as career needs.
At a structural level, the boundaries between organisations, professions and the health and social care sectors may need to be addressed as potential barriers to successful innovation.
More research is needed to confirm whether addressing these factors prospectively enhances the success of future innovations.
Supplementary Material
Acknowledgments.
We would like to thank Josh Brewster from the Health Innovation Network and Sian Kitchen from Health Education England for their commitment to this project, and Professor Sue Smith from the Medical Education Research Unit of Imperial College for ongoing support and advice.
Twitter: @doctorkayleigh
Contributors: KL-G and GBR contributed to the conception and design of the work and to the acquisition of data. KL-G, GBR and AK collaborated on the data analysis and interpretation. KL-G and AK drafted the work, all authors revised it critically for important intellectual content. All authors have approved the final version and agree to be accountable for all aspects of the work and to resolve questions relating to accuracy or integrity.
Funding: This work was funded by Health Education England (grant number XXMLIVESEY).
Disclaimer: Gabriel Reedy is affiliated to the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Emergency Preparedness and Response at King’s College London in partnership with Public Health England (PHE), in collaboration with the University of East Anglia and Newcastle University. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or Public Health England. These funders had no input in the writing of and the decision to submit this article.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Ethics statements, patient consent for publication.
Not required.
Ethics approval
Ethical approval was granted on 26 March 2019 by the Research Ethics Committee of King’s College London (LRS-18/19-10432).
- Publications
- News and Events
- Education and Outreach
Software Engineering Institute
The Critical Success Factor Method: Establishing a Foundation for Enterprise Security Management
July 1, 2004 • technical report, by richard a. caralli , james f. stevens , bradford j. willke , and william r. wilson, cmu/sei report number, doi (digital object identifier), topic or tag.
Every organization has a mission that describes why it exists (its purpose) and where it intends to go (its direction). The mission reflects the organization's unique values and vision. Achieving the mission takes the participation and skill of the entire organization. The goals and objectives of every staff member must be aimed toward the mission. However, achieving goals and objectives is not enough. The organization must perform well in key areas on a consistent basis to achieve the mission. These key areas—unique to the organization and the industry in which it competes—can be defined as the organization's critical success factors.
The critical success factor method is a means for identifying these important elements of success. It was originally developed to align information technology planning with the strategic direction of an organization. However, in research and fieldwork undertaken by members of the Survivable Enterprise Management (SEM) team at the Software Engineering Institute, it has shown promise in helping organizations guide, direct, and prioritize their activities for developing security strategies and managing security across their enterprises. This report describes the critical success factor method and presents the SEM team's theories and experience in applying it to enterprise security management.
Cite This Technical Report
Caralli, R., Stevens, J., Willke, B., & Wilson, W. (2004, July 1). The Critical Success Factor Method: Establishing a Foundation for Enterprise Security Management. (Technical Report CMU/SEI-2004-TR-010). Retrieved September 1, 2024, from https://doi.org/10.1184/R1/6585107.v1.
@techreport{caralli_2004, author={Caralli, Richard and Stevens, James and Willke, Bradford and Wilson, William}, title={The Critical Success Factor Method: Establishing a Foundation for Enterprise Security Management}, month={Jul}, year={2004}, number={CMU/SEI-2004-TR-010}, howpublished={Carnegie Mellon University, Software Engineering Institute's Digital Library}, url={https://doi.org/10.1184/R1/6585107.v1}, note={Accessed: 2024-Sep-1} }
Caralli, Richard, James Stevens, Bradford Willke, and William Wilson. "The Critical Success Factor Method: Establishing a Foundation for Enterprise Security Management." (CMU/SEI-2004-TR-010). Carnegie Mellon University, Software Engineering Institute's Digital Library . Software Engineering Institute, July 1, 2004. https://doi.org/10.1184/R1/6585107.v1.
R. Caralli, J. Stevens, B. Willke, and W. Wilson, "The Critical Success Factor Method: Establishing a Foundation for Enterprise Security Management," Carnegie Mellon University, Software Engineering Institute's Digital Library . Software Engineering Institute, Technical Report CMU/SEI-2004-TR-010, 1-Jul-2004 [Online]. Available: https://doi.org/10.1184/R1/6585107.v1. [Accessed: 1-Sep-2024].
Caralli, Richard, James Stevens, Bradford Willke, and William Wilson. "The Critical Success Factor Method: Establishing a Foundation for Enterprise Security Management." (Technical Report CMU/SEI-2004-TR-010). Carnegie Mellon University, Software Engineering Institute's Digital Library , Software Engineering Institute, 1 Jul. 2004. https://doi.org/10.1184/R1/6585107.v1. Accessed 1 Sep. 2024.
Caralli, Richard; Stevens, James; Willke, Bradford; & Wilson, William. The Critical Success Factor Method: Establishing a Foundation for Enterprise Security Management . CMU/SEI-2004-TR-010. Software Engineering Institute. 2004. https://doi.org/10.1184/R1/6585107.v1
- Home > B2B Blog > 6 Critical Success Factors of a Qualitative Research Project
6 Critical Success Factors of a Qualitative Research Project

Over the last couple of years qualitative research has made a comeback and piqued the interest of many of our clients in the b2b sector. In taking the lead for qual research here at B2B International HQ, I’ve compiled a list of the 6 critical factors for delivering a successful qualitative research project.
Using the Right Methodology
The crux of any research project is using the correct methodology but within qualitative research it can be even more important. In order to choose the right methodology you first need to understand your research objectives. Once you know what you want to find out, you need to ask yourself one question:
“Does this topic require group discussion or individual in-depth understanding?”
In answering this question you then have a basis for whether your methodology is based around focus groups or in-depth interviews (I’m including ethnography in this for simplicity!).
As an example, in-depth interviews (IDIs) will be more appropriate to gather an intricate understanding of a decision making unit whereas focus groups will be better for discussing a new creative concept.
Talking To the Right Person
With a much smaller sample size with a need to gather greater depth, it’s imperative that the respondent is knowledgeable about the subject and, in b2b markets, that they have significant influence within their company on the subject.
Compiling your list is going to play a huge part in this and in some instances this may need some troubleshooting; particularly if you aren’t 100% sure on who you need to be talking to.
Briefing Interviewers & Moderators
When briefing your interviewers and/or moderators (where necessary) it’s important to remember to get across the main research objective as THE most critical piece of information, rather than focusing on the small details.
A good interviewer or moderator will be able to bring out the key themes from knowing the high-level project objectives.
Note Taking or Transcripts
This one is an area of personal preference but whatever your methodology you will need at least one of them!
The important thing to note here is that when taking notes at a focus group or IDI you must write verbatim (as much as possible) without analysing what’s being said. It should be left for you or the person analysing the notes afterwards to make that judgement.
Reading Transcripts
Transcripts from IDIs should be read as single entities first to get a ‘feel’ for each interview i.e. things that aren’t actually written down. (This is also something you can ask the interviewer to do after each interview).
It is good practice not to leave all this to the end of fieldwork either! You will have a lot of data to read through so it should be done as you go along.
Qualitative Analysis
This type of data should be treated very differently to quantitative data, in most cases you aren’t looking for percentages but instead want themes.
Digging into your data, you should look for themes which can be linked to categories and eventually frameworks. There are also a number of different techniques we can utilise during analysis including Grounded Theory, Interpretive Phenomenological Analysis and Discourse Analysis. (Look out for my next blogs which will go into these techniques in detail!)
Learn More About Qualitative Research >
Contact Us >
To learn more about our full range of research services and how they can help your business
Recent Articles
- Derived Importance: What Is It, and Why Does It Matter?
- Navigating Remote Training: Turning Challenges into Opportunities
- The Benefits of Conducting In-Person Focus Groups
- The Importance of Active Listening in Business Communication and Market Research
- When to Use Qualitative Research to Better Understand Your Customers and Their Needs
Speak to an expert...
- Keep up to date with our latest research
- First name *
- Last name *
- Company name *
- By subscribing to this newsletter, you are opting in to our marketing communications. Read our privacy policy for more information.
- Phone This field is for validation purposes and should be left unchanged.
Privacy Overview
| Cookie | Duration | Description |
|---|---|---|
| __hssrc | session | This cookie is set by Hubspot whenever it changes the session cookie. The __hssrc cookie set to 1 indicates that the user has restarted the browser, and if the cookie does not exist, it is assumed to be a new session. |
| cookielawinfo-checkbox-advertisement | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to record the user consent for the cookies in the "Advertisement" category . |
| cookielawinfo-checkbox-analytics | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Analytics". |
| cookielawinfo-checkbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-others | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Other. |
| cookielawinfo-checkbox-performance | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Performance". |
| CookieLawInfoConsent | 1 year | Records the default button state of the corresponding category & the status of CCPA. It works only in coordination with the primary cookie. |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
| Cookie | Duration | Description |
|---|---|---|
| __cf_bm | 30 minutes | This cookie, set by Cloudflare, is used to support Cloudflare Bot Management. |
| __hssc | 30 minutes | HubSpot sets this cookie to keep track of sessions and to determine if HubSpot should increment the session number and timestamps in the __hstc cookie. |
| bcookie | 2 years | LinkedIn sets this cookie from LinkedIn share buttons and ad tags to recognize browser ID. |
| bscookie | 2 years | LinkedIn sets this cookie to store performed actions on the website. |
| lang | session | LinkedIn sets this cookie to remember a user's language setting. |
| lidc | 1 day | LinkedIn sets the lidc cookie to facilitate data center selection. |
| UserMatchHistory | 1 month | LinkedIn sets this cookie for LinkedIn Ads ID syncing. |
| Cookie | Duration | Description |
|---|---|---|
| __hstc | 5 months 27 days | This is the main cookie set by Hubspot, for tracking visitors. It contains the domain, initial timestamp (first visit), last timestamp (last visit), current timestamp (this visit), and session number (increments for each subsequent session). |
| _ga | 2 years | The _ga cookie, installed by Google Analytics, calculates visitor, session and campaign data and also keeps track of site usage for the site's analytics report. The cookie stores information anonymously and assigns a randomly generated number to recognize unique visitors. |
| _gat_gtag_UA_3031018_3 | 1 minute | Set by Google to distinguish users. |
| _gid | 1 day | Installed by Google Analytics, _gid cookie stores information on how visitors use a website, while also creating an analytics report of the website's performance. Some of the data that are collected include the number of visitors, their source, and the pages they visit anonymously. |
| CONSENT | 2 years | YouTube sets this cookie via embedded youtube-videos and registers anonymous statistical data. |
| hubspotutk | 5 months 27 days | HubSpot sets this cookie to keep track of the visitors to the website. This cookie is passed to HubSpot on form submission and used when deduplicating contacts. |
| undefined | never | Wistia sets this cookie to collect data on visitor interaction with the website's video-content, to make the website's video-content more relevant for the visitor. |
| vuid | 2 years | Vimeo installs this cookie to collect tracking information by setting a unique ID to embed videos to the website. |
| Cookie | Duration | Description |
|---|---|---|
| VISITOR_INFO1_LIVE | 5 months 27 days | A cookie set by YouTube to measure bandwidth that determines whether the user gets the new or old player interface. |
| YSC | session | YSC cookie is set by Youtube and is used to track the views of embedded videos on Youtube pages. |
| yt-remote-connected-devices | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| yt-remote-device-id | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| Cookie | Duration | Description |
|---|---|---|
| AnalyticsSyncHistory | 1 month | No description |
| closest_office_location | 1 month | No description |
| li_gc | 2 years | No description |
| loglevel | never | No description available. |
| ssi--lastInteraction | 10 minutes | This cookie is used for storing the date of last secure session the visitor had when visiting the site. |
| ssi--sessionId | 1 year | This cookie is used for storing the session ID which helps in reusing the one the visitor had already used. |
| user_country | 1 month | No description available. |
- Search Menu
- Sign in through your institution
- Chemical Biology and Nucleic Acid Chemistry
- Computational Biology
- Critical Reviews and Perspectives
- Data Resources and Analyses
- Gene Regulation, Chromatin and Epigenetics
- Genome Integrity, Repair and Replication
- Nucleic Acid Enzymes
- RNA and RNA-protein complexes
- Synthetic Biology and Bioengineering
- Molecular and Structural Biology
- Advance Articles
- Breakthrough Articles
- Molecular Biology Database Collection
- Special Collections
- Scope and Criteria for Consideration
- Author Guidelines
- Data Deposition Policy
- Database Issue Guidelines
- Web Server Issue Guidelines
- Submission Site
- About Nucleic Acids Research
- Editors & Editorial Board
- Information of Referees
- Self-Archiving Policy
- Dispatch Dates
- Advertising and Corporate Services
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic
Article Contents
Introduction, materials and methods, data availability, supplementary data, acknowledgements, identification of transcription factor co-binding patterns with non-negative matrix factorization.
- Article contents
- Figures & tables
- Supplementary Data
Ieva Rauluseviciute, Timothée Launay, Guido Barzaghi, Sarvesh Nikumbh, Boris Lenhard, Arnaud Regis Krebs, Jaime A Castro-Mondragon, Anthony Mathelier, Identification of transcription factor co-binding patterns with non-negative matrix factorization, Nucleic Acids Research , 2024;, gkae743, https://doi.org/10.1093/nar/gkae743
- Permissions Icon Permissions
Transcription factor (TF) binding to DNA is critical to transcription regulation. Although the binding properties of numerous individual TFs are well-documented, a more detailed comprehension of how TFs interact cooperatively with DNA is required. We present COBIND, a novel method based on non-negative matrix factorization (NMF) to identify TF co-binding patterns automatically. COBIND applies NMF to one-hot encoded regions flanking known TF binding sites (TFBSs) to pinpoint enriched DNA patterns at fixed distances. We applied COBIND to 5699 TFBS datasets from UniBind for 401 TFs in seven species. The method uncovered already established co-binding patterns and new co-binding configurations not yet reported in the literature and inferred through motif similarity and protein-protein interaction knowledge. Our extensive analyses across species revealed that 67% of the TFs shared a co-binding motif with other TFs from the same structural family. The co-binding patterns captured by COBIND are likely functionally relevant as they harbor higher evolutionarily conservation than isolated TFBSs. Open chromatin data from matching human cell lines further supported the co-binding predictions. Finally, we used single-molecule footprinting data from mouse embryonic stem cells to confirm that the COBIND-predicted co-binding events associated with some TFs likely occurred on the same DNA molecules.

The interactions between DNA and transcription factors (TFs) are crucial to transcription regulation as they intrinsically determine cell growth, development, and response to stimuli. TFs bind DNA at TF binding sites (TFBSs) in a sequence-specific manner through direct contact between DNA nucleotides and amino acids of the TFs’ DNA-binding domain (DBD) ( 1 ). Structural similarities between DBDs classify TFs into structural classes and families, and TFs from the same class or family usually recognize similar DNA patterns (or motifs).
The binding properties of individual TFs have been vastly studied ( 2–4 ), and several databases store DNA binding profiles for TFs across multiple taxonomic groups (e.g. JASPAR, CIS-BP and HOCOMOCO ( 5–7 )). While these databases primarily provide TF binding motifs for individual TFs, there is a need to increase our understanding of how TFs cooperatively bind DNA to regulate transcription ( 8 ). The cooperative binding of TFs generates many possible binding combinations, thus increasing the complexity of gene regulatory networks ( 8 , 9 ). Recent studies argue for a flexible grammar of motifs at cis- regulatory regions, where the spacing and orientation between TFBSs would not be a key determinant for transcription regulation ( 10–13 ). Nevertheless, some TFs physically cooperate, providing a strict motif syntax important for transcription regulation. For instance, exhaustively testing the combinatorics of liver-associated TFBSs with massively parallel reporter assays revealed that TFBS orientation and order are major drivers of gene regulatory activity ( 14 ). A well-characterized example of physical interaction between TFs is POU5F1 (OCT4) cooperation with either SOX2 or SOX17 in pluripotent cells. The spacing between the binding sites will determine POU5F1’s partner and the corresponding regulatory effect ( 15 , 16 ). Hence, the specific spacing and orientation between the canonical motifs recognized by two TFs can characterize their co-binding at given genomic regions ( 17 , 18 ). However, the cooperative binding of two TFs can also slightly modify their individual DNA sequence preference ( 19 ). Consequently, systematically identifying cooperative events genome-wide with strict binding grammar is still challenging.
The CAP-SELEX experimental technique captures co-binding events between predefined sets of TFs in vitro ( 19 ). However, whether the same co-binding properties will occur in vivo remains to be determined. Therefore, computational methods, such as TF-COMB, TACO, SpaMo and MCOT ( 20–23 ), leverage in vivo data, such as ChIP-seq ( 24 ), to predict co-binding events. These tools rely on identifying the co-occurrence of pre-defined genomic regions bound by the individual TFs or already known individual DNA binding motifs in predefined genomic regions. While these strategies reduce the search space, they restrict discoveries for the pairs of TFs where either bound regions or TF binding motifs exist for both TFs. When relying on already-known TF binding motifs, the quality of the available motif collections is a limiting factor, and this approach cannot discover new DNA sequence patterns. Another approach implemented in the RSAT dyad-analysis tool does not rely on pre-defined motifs and predicts spaced pairs of motifs de novo starting from spaced 3-mer patterns ( 25 , 26 ). Finally, deep learning approaches can infer regulatory patterns from experimental data, with the capacity to pinpoint TF co-binding events ( 12 , 27 ). Even though the underlying algorithms are advanced and powerful, their complexity and interpretability make it challenging to characterize specific co-binding events without extensive a posteriori analyses.
In this study, we aimed to discover TF co-binding patterns in the vicinity of high-quality TFBSs (referred as anchors). The discovery of fixed co-binding patterns can be considered a matrix decomposition (or factorization) problem where an input matrix encodes nucleotides surrounding TFBSs. The ultimate goal is to group nucleotide patterns from this matrix. The non-negative matrix factorization (NMF) technique decomposes a given non-negative matrix into two low-rank, non-negative matrices to reveal underlying patterns and structures within the data ( 28 , 29 ). This technique has been useful in computational biology to reveal molecular patterns from high-throughput data ( 28 , 30 ). seqArchR is the first novel application of NMF for the simultaneous identification of sequence features and corresponding clusters ( 31 ). The seqArchR tool identifies critical DNA elements in promoter regions by applying NMF to the corresponding one-hot encoded sequences ( 31 ). This approach inspired us to use NMF to address the discovery of TF co-binding patterns.
We present COBIND, a Snakemake-based ( 32 ) pipeline for the de novo discovery of TF co-binding patterns from input sets of TFBSs ( https://bitbucket.org/CBGR/cobind_tool ). We applied COBIND to 5699 sets of high-quality TFBSs from seven species ( Arabidopsis thaliana , Caenorhabditis elegans , Danio rerio , Drosophila melanogaster , Homo sapiens , Mus musculus and Rattus norvegicus ) for 401 unique TFs stored in the UniBind database ( 33 ). COBIND recovered established and unreported co-binding events between TFs. Among TFs from the same structural family, the majority shared co-binding patterns. In human and mouse genomes, genomic regions harboring the predicted co-binding events are evolutionarily more conserved than genomic regions without co-binding. Moreover, increased chromatin accessibility in matching human cell lines supported the predicted co-binding events from bulk experimental data. Finally, using single-molecule footprinting data from mouse embryonic stem cells, we confirmed that the anchor and co-binding patterns associated with some TFs are significantly enriched for TF co-occupancy at the single-molecule level. Overall, COBIND can de novo discover regions in the genomes that harbor patterns of co-binding events between TFs.
Predicting co-binding patterns with COBIND
COBIND takes a BED (or FASTA) formatted file providing the genomic coordinates of anchor regions (or DNA sequences centered at the anchor sites), such as TFBS locations, as input. The tool uses NMF to reveal recurring DNA motifs with space constraints in the regions surrounding the input anchors, which are not factorized. The underlying computational pipeline consists of the following steps (Figure 1 ):
Step 1: Extraction of flanking regions . COBIND extracts the DNA sequences surrounding the anchor TFBSs ( n bp upstream and downstream; n = 30 by default) using bedtools (v2.29.2) ( 34 ).
Step 2: One-hot encoding . The flanking sequences are one-hot encoded as vectors of 4 bits per DNA nucleotide (A: 1000, C: 0100, G: 0010, T: 0001; ambiguous nucleotides |$ \notin [ {A,\ C,\ G,\ T} ]$| are encoded as 1111 (Figure 1 Step 2)). COBIND constructs two matrices representing the upstream and downstream sequences by combining the vectors of one-hot encoded sequences. Each row corresponds to a sequence flanking an anchor TFBS from the input set.
Step 3: Non-negative matrix factorization . COBIND applies NMF to each matrix using the NMF function of scikit-learn (v0.23.2) ( 35 ). The NMF decomposes an input matrix into two matrices: one representing k components (or factors) of nucleotide patterns and one informing the presence of the identified patterns in each row of the input matrix ( 28 , 30 ). Importantly, COBIND applies NMF with multiple values of k to increase its capacity to capture co-binding patterns with different resolutions (see section ‘Parameter settings’ below) (Figure 1 Step 3). Next, COBIND assigns input sequences to each component following the methodology described by Kim and Park ( 36 ). Finally, COBIND builds motifs as positional frequency matrices (PFM) for each component by computing the occurrence frequencies of each nucleotide at each position (Additional file 1: Supplementary Figure S1 ).
Step 4: Motifs filtering . COBIND filters out PFMs with information content (IC) < 2 to focus on informative patterns. Next, it aims to identify motifs corresponding to positions with local enrichment of high IC to distinguish them from motifs with scattered high IC positions or equal distribution of IC along the flanking region (Additional file 1: Supplementary Figure S1 ). Specifically, COBIND computes the Gini coefficient g of each PFM to measure the inequality of IC values ( 37 ). We discard PFMs with a low Gini coefficient ( g < 0.5 by default).
Step 5: Motif trimming . COBIND extracts the positions corresponding to the local enrichment of high IC values, revealing co-binding motifs. Specifically, it first computes the smallest window of size n that contains at least half of the IC of the complete flank. Finally, COBIND doubles the size of the window (adding n /2 nucleotides up and downstream) to define the co-binding motif.
Step 6: Motif clustering . COBIND can identify similar motifs multiple times since (i) NMF runs with different values for k , (ii) similar motifs can have different spacings with the core motif, and (iii) multiple TFBS sets can be processed in parallel, resulting in the identification of similar motifs (Figure 1 Step 3). COBIND provides non-redundant motifs by clustering the identified motifs (Figure 1 Step 6). Specifically, we integrate the RSAT matrix-clustering tool ( 38 ) to cluster similar motifs (we provide the used parameters in Additional file 1: Supplementary Table S1 ). For each motif cluster, COBIND constructs an archetypal motif following the methodology from ( 39 ), which summarizes motifs by computing the means of the counts from the aligned matrices in each cluster. We discarded successive flanking positions with IC < 0.1 from the archetypal motif.
Similarly, we cluster and summarize the anchor TFBS sequences associated with each co-binding motif (Additional file 1: Supplementary Table S1 ).

Schematic workflow of COBIND. 1) COBIND takes one or multiple input file(s) that provide regions of interest, such as TFBSs. The tool extracts upstream and downstream DNA sequences (default: 30 nt each). 2) The sequences are one-hot encoded to construct encoded matrices of upstream and downstream sequences. 3) COBIND applies NMF on each matrix to extract DNA patterns. 4) The tool computes the Gini coefficient of each DNA pattern and discards the uninformative patterns. 5) COBIND trims the identified flanking patterns to reveal possible TF co-binding motifs. 6) RSAT matrix-clustering groups redundant motifs, and COBIND constructs archetypal motifs summarizing each cluster of similar motifs. 7) The predicted co-binding patterns are visualized through a heatmap representing the predicted spacing between the anchor (at the center) and the co-binding patterns with the corresponding orientation. The predicted co-binding patterns are provided on the left and/or right of the plots, depending on whether they were found upstream and/or downstream of the anchor sites. The position of the heatmap tiles provides, for each pattern, the number of nucleotides between the co-binding and anchor sites. The color of a heatmap tile represents the number of input datasets in which the co-binding pattern was predicted. The dot size and number within the heatmap tileblocks correspond to the proportion of input sequences in which a co-binding instance with a specific spacing was predicted.
Step 7: Summarize the discovered co-binding events . COBIND computes the relative spacing and orientation to the anchor motif from each co-binding archetypal motif discovered. Finally, it illustrates the corresponding co-binding events as heatmaps (Figure 1 Step 7). The spacing summary plot describes the relationship between the anchor (provided at the center of the plot) and the discovered co-binding patterns (upstream or downstream from the anchor). On the x-axis, one visualizes the number of nucleotides between the anchor and the discovered co-binding pattern motifs, illustrated as sequence logos. The color intensity in the heatmap illustrates the proportion of the input datasets with predicted co-binding motif instances. The inserted number indicates the proportion of unique sequences in the datasets with the corresponding co-binding pattern, orientation, and spacing, that are predicted to harbor the corresponding co-binding configurations.
Parameter settings
The hyperparameters of COBIND are the length of the flanking regions, the number of components for the different runs of the NMF, and the Gini coefficient threshold. Below, we describe the values used in this study for each parameter and why we selected these values.
Length of the flanking sequences . We considered 30 bp upstream and downstream of the input TFBSs to report co-binding events likely associated with physical interactions between the TFs. The longest TFBS motif used in UniBind is about 20 bp-long ( 33 ), and cooperative TFs bind TFBSs 9–10 bp away from each other on average ( 19 ). Furthermore, larger flanking regions resulted in noisier co-binding motifs and could miss co-binding motif configurations (Additional file 1: Supplementary Figure S2 ).
Number of components . COBIND runs NMF several times with different numbers of components ( k ). We used synthetic data to estimate a suitable range of values for k . Specifically, we implanted different proportions of instances of predefined motifs in random sequences. We considered sets of 800, 1000, 4000, 8000, 10 000, 40 000 and 80 000 synthetic sequences. For each set of synthetic sequences, we injected an instance of the considered motif in 0.5, 1, 3, 5 and 10% of the sequences. We ran COBIND with k ∈ [2, 17] for each synthetic dataset. For each k value, we evaluated specificity and sensitivity by computing the F1 score and Matthew's correlation coefficient (MCC) for each discovered motif. Moreover, we reported the proportion of correctly predicted motifs. We considered that COBIND predicted the correct motif if it was similar to the injected motif—as assessed by Tomtom ( 40 ) with a P -value <0.05 (Additional file 1: Supplementary Table S1 ). We used the results from the synthetic data to select k ∈ [3, 6] for further analyses (Additional file 1: Supplementary Figures S3 – S15 ).
Gini coefficient threshold . To empirically determine the Gini coefficient threshold, we compared the distributions of Gini coefficients for the motifs predicted by COBIND on the TFBS datasets from UniBind (see below) with those of predictions from random sequences. For each UniBind set of TFBSs, we constructed a set of synthetic sequences by shuffling the DNA sequences flanking the TFBSs with the kmer shuffling module of the BiasAway tool with k = 1 ( 41 ). For each species considered, we selected the Gini coefficient threshold corresponding to a 1% false discovery rate (Additional file 1: Supplementary Figure S16 ). This strategy resulted in the following Gini coefficient thresholds: 0.50 for Arabidopsis thaliana , 0.46 for Caenorhabditis elegans , 0.48 for Danio rerio , 0.49 for Drosophila melanogaster , 0.50 for Homo sapiens , 0.54 for Mus musculus and 0.53 for Rattus norvegicus . Finally, we estimated the stability of the thresholds obtained with this strategy by performing the same analysis multiple times and subsampling the number of datasets from H. sapiens and M. musculus . The Gini coefficient thresholds were stable across the different runs (Additional file 1: Supplementary Figure S17 ).
Transcription factor binding site datasets
We used the robust collection of TFBS sets from the UniBind database (2021 version) ( https://unibind.uio.no/ ) derived from 7 species ( Arabidopsis thaliana , Caenorhabditis elegans , Danio rerio , Drosophila melanogaster , Homo sapiens , Mus musculus, Rattus norvegicus ) ( 33 ). UniBind TFBS datasets derive from pairs of ChIP-seq peak datasets and JASPAR TF binding profiles. When multiple TF binding profiles exist for a given TF, we selected, for each corresponding ChIP-seq dataset, the profile providing the best centrality P -value as assessed in UniBind. We did not consider JASPAR TF binding profiles associated with TF dimers. Datasets with <1000 TFBSs were filtered out (Additional file 1: Supplementary Table S2 ).
Benchmarking
Synthetic datasets.
We generated the synthetic datasets following the methodology mentioned above. We constructed synthetic datasets with 800, 1000, 4000, 8000, 10 000, 40 000 and 80 000 sequences with instances of a known motif inserted in 0.5, 1, 3, 5 and 10% of the sequences. We considered 13 motifs (3- to 19-bp long) from the JASPAR database ( 7 ), which were synthetically inserted into random sequences with varying spacing (6- to 22-bp) from the anchor sites. To run SpaMO, we inserted the CTCF (MA0139.1) motif at the center of the flanking sequences since SpaMO requires a motif to anchor its search for co-binding patterns.
Comparisons between COBIND and other tools
We used the synthetic data to compare COBIND with STREME ( 42 ), RSAT peak-motifs dyad-analysis ( 25 ), RSAT peak-motifs oligo-analysis ( 26 , 43 , 44 ), RSAT peak-motifs position-analysis ( 43 , 44 ) and SpaMo ( 21 ). We compared the motifs predicted by the different tools with the original inserted motifs using Tomtom ( 40 ) (Additional file 1: Supplementary Table S1 ). We considered a discovered motif as a match to the injected motif if Tomtom predicted them to be similar with a P -value <0.05. We provide the number of incorrectly predicted motifs. To run SpaMo, we used the JASPAR 2020 CORE collection of non-redundant profiles ( 45 ). We computed the F1 score and Matthew's correlation coefficient (MCC) to assess the capacity of the tools to retrieve the correct sequences where the motifs were injected. When the correct motif was predicted multiple times, we reported the one providing the highest F 1 score.
STREME and RSAT algorithms were run to discover a maximum of 18 motifs to match the number of possible motifs to be predicted by COBIND. All tools were run with default parameters otherwise. When estimating the running time, we used the parallelizable version of the NMF for COBIND.
Inference of the co-binding transcription factors from the discovered co-binding motifs
We assessed the similarities between a predicted co-binding motif and known motifs associated with 5272 TFs—collected from JASPAR 2022 non-redundant CORE, JASPAR 2022 non-redundant Unvalidated taxon-specific, and CIS-BP ( 5 )—using Tomtom (Additional file 1: Supplementary Table S1 ) ( 40 ). We predicted TF B to bind the predicted co-binding motif associated with anchor TFBSs of TF A if Tomtom reported a similarity P -value <0.05 with the known canonical motif bound by TF B (criteria I). To assess possible physical interactions between TF A and TF B , we retrieved protein-protein interaction (PPI) data from STRING v11.0 ( 46 ). We considered TF A and TF B to interact physically when their STRING PPI score was above or equal to 500 (criteria II). We reported the co-binding pair TF A -TF B to be already ‘known’ when they met criteria I and II. We reported TF A –TF B as a novel co-binding pair when they met criteria I but not II.
Shared co-binding motifs across transcription factor structural families
To assess the conservation of co-binding motifs across TFs, we ran COBIND (up to Step 5) on all UniBind datasets. We independently clustered all the predictions (COBIND Step 6) for each TF structural family; note that this step considered TFs from multiple species. TFs without known structural families were excluded from this analysis. When the co-binding motifs associated with two (or more) TFs from the same family were similar (i.e. belonged to the same cluster), we considered the TFs to share the same co-binding motif.
Evolutionary conservation
We downloaded the following conservation tracks in bigwig format from the UCSC genome browser: phastCons100way (human; hg38), phastCons60way (mouse; mm10), phastCons135way ( C. elegans ; WBcel235), and phastCons27way ( D. melanogaster ; dm6) ( 47 ). We obtained the 63 flowering plants PhastCons conservation track from PlantRegMap ( 48 ) for analyzing datasets from A. thaliana . We computed the conservation scores of a genomic region using the aggregate and extract functions of bwtool (v1.0) ( 49 ).
We compared the distributions of conservation scores between two regions with one-sided Wilcoxon signed-rank tests. Specifically, we compared the conservation values at the anchor (or co-binding) sites between the genomic regions predicted to harbor a co-binding pattern or not. These comparisons result in two P -values per co-binding pattern - one comparing the anchor sites and one comparing the co-binding sites. Histograms were used to summarize the P -values across datasets, considering the co-binding pattern with the lowest P -value per dataset. We compared the P -values to random expectation. Specifically, we randomly assigned, for each dataset, each genomic region to harbor or not a co-binding pattern (keeping the same number of regions in each category as the number predicted by COBIND). We performed five randomizations per dataset and reported the combined results.
DNase I hypersensitive footprinting analysis
We downloaded DNase I hypersensitivity (DHS) data generated by the ENCODE consortium for multiple human cell types in bigwig format at https://resources.altius.org/∼jvierstra/projects/footprinting.2020/ ( 50 ). We considered the 53 cell lines or tissues for which both DHS and COBIND predictions were available; we analyzed 66 DHS datasets associated with 70 co-binding configurations for 44 TF binding profiles as anchors. We followed the methodology described above to compare evolutionary conservation scores when comparing the DHS signal between two genomic regions (with or without a co-binding event predicted by COBIND).
Single-molecule footprinting analysis
We downloaded 12 double enzyme single-molecule footprinting (SMF) datasets produced from triple DNA methyltransferases knockout lines of mouse embryonic stem cells (mESCs) from ArrayExpress with accession numbers E-MTAB-9033 and E-MTAB-9123 ( 51 ). Whenever applicable, we performed the analyses of SMF data with the SingleMoleculeFootprinting R package following the instructions previously described ( 52 ). To compare the SMF data in the genomic regions predicted by COBIND to contain or not a co-binding event in mESC datasets, we modified the SortReadsByTFCluster_MultiSiteWrapper functions from the development version of the SingleMoleculeFootprinting package ( 53 ) ( https://bitbucket.org/CBGR/cobind_manuscript/src/master/bin/single_molecule/analyse_sm/sort_tf_reads_by_cobind_clusters.R ). Using the methylation status provided by the SMF data, we determined each genomic region to be either (i) free of nucleosomes (‘Accessible’), (ii) occupied by nucleosomes (‘Nucleosome’), (iii) bound only by the anchor (‘Anchor’), (iv) bound only by the co-binding TF (‘Co-binding’) or (v) bound by both the anchor and co-binding TF (‘Anchor + Co-binding’). We summed the number of molecules for each state in different samples and computed the proportions of molecules. We summarized the proportions of each state for all predictions and all anchors together. We compared the proportions of each state in genomic regions predicted to contain a co-binding event or not using a two-sided Wilcoxon signed-rank test. We compared the same type of regions in the ‘Anchor + Co-binding’ state for individual anchors using a one-sided Wilcoxon signed-rank test.
COBIND discovers co-binding patterns de novo
Comparisons with other tools on simulated data.
We report a new computational framework, COBIND, to predict space-fixed co-binding patterns in the vicinity of TFBSs provided as input. COBIND relies on applying NMF to the one-hot encoded regions flanking the user-provided TFBSs to predict co-occurring DNA motifs with fixed spacing (Materials and methods). We compared COBIND to other tools discovering motifs de novo (STREME, RSAT dyad-analysis , RSAT oligo-analysis and RSAT position-analysis ). Additionally, we compared COBIND to SpaMO, which predicts spatially co-occurring instances of known motifs (Additional file 1: Supplementary Table S3 ). For comparisons, we generated synthetic data by injecting instances of known motifs (13 distinct motifs were used) at different frequencies in random sets of 800 to 80 000 DNA sequences (Materials and methods).
COBIND predicted the correct motif inserted when considering a minimum of 800 sequences for 10 of the 13 used inserted motifs. For example, COBIND accurately predicted the injected ATF4 motif in datasets of varying sizes without incorrect motif predictions (Figure 2 ). When considering the shorter TAL1 motif (3-bp known to be co-bound by the dimer GATA:TAL1), COBIND provides the most accurate predictions in multiple datasets when other tools obtain lower F 1 scores and MCC values and predict incorrect motifs (Additional file 1: Supplementary Figure S18 - S29 ). Overall, COBIND predicted no false positive motif across most configurations (see the number of incorrect motifs discovered in Additional file 1: Supplementary Figure S18 - S29 ). In most cases, COBIND discovered the injected motifs in the correct sequences, as illustrated by high F 1 scores and MCC values. Comparably, STREME discovered only correct motifs but failed to predict any motifs in some configurations (especially when the number of sequences with the injected motif was low). When considering many sequences (datasets of 40 000 and 80 000 sequences), COBIND and STREME recovered the correct motifs. SpaMo discovered the correct inserted motif in most cases, but it came at the cost of predicting many false positives. One should note that SpaMo requires a reference set of known motifs to predict co-occurrences, while COBIND predicts co-binding motifs de novo .

Comparisons between COBIND and other tools on simulated data. We ran COBIND, STREME, RSAT-dyads, RSAT-oligos, RSAT-positions and SPAMO (subfacets) on simulated data containing different numbers of sequences (from 800 to 80 000, see facets) with inserted JASPAR MA0833.2 TF binding motif instances for ATF4 (see Methods for details). We considered (i) F 1 scores and Matthew's correlation coefficient (MCC) (in case multiple correct motifs are found, a motif with the highest F 1 score is visualized). (ii) the number of incorrect motifs discovered in each experiment. Good performance of a tool would be indicated by a discovered motif with a high F 1 score and high MCC and no incorrectly discovered motifs.
COBIND exhibited a similar run time to the other tools on datasets with up to 10 000 sequences. Nevertheless, it was slower than the other tools, except STREME, with larger datasets (>40 000 sequences) due to the motif clustering step applied to the results of the NMF with 3–6 components. Notwithstanding, the COBIND workflow (from motif discovery with NMF to co-binding summary) takes ∼500 s on 40 000 sequences as it allows for parallelization across CPU cores (Additional file 1: Supplementary Figure S30 ).
Proof-of-concept: POU5F1 co-binding with SOX2 or SOX17
As a proof-of-concept, we applied COBIND to UniBind TFBS datasets for the SOX2 and SOX17 TFs. Previous studies established that both TFs partner with POU5F1 in pluripotent cells, where POU5F1 co-binds with SOX2 at regulatory elements to promote pluripotency. At the same time, it co-binds with SOX17 to control endoderm differentiation at other regulatory elements ( 11 , 16 ). While POU5F1 and SOX2 cooperate through binding at instances of their respective canonical motifs, POU5F1 can associate with SOX17 to bind a compressed motif (Figure 3A ). COBIND successfully discovered the two co-binding patterns - canonical and compressed - recognized by POU5F1 when applied to the SOX2 and SOX17 TFBS datasets from H. sapiens and M. musculus (Figure 3B ). COBIND retrieved the correct pattern in 11 human SOX2 datasets (58%), with ∼7% of the sequences predicted to harbor the pattern, and in 20 mouse datasets (27%), with ∼6% of the sequences harboring the pattern (Additional file 1: Supplementary Figure S31A, B ). In agreement with previous studies, the datasets where COBIND predicted the pattern derived from human and mouse ESC and mouse embryonic fibroblast ( 16 , 18 , 54 ). When considering the SOX17 datasets, COBIND successfully discovered the canonical co-binding pattern and the compressed motif. It predicted the compressed co-binding pattern in two datasets (50%; derived from mouse ESC), with 5% of the sequences harboring the co-binding motif (Additional file 1: Supplementary Figure S31C ).

Application of COBIND to SOX2 and SOX17 TFBS datasets. COBIND predicts POU5F1 to co-bind with SOX2 (human and mouse) and SOX17 (mouse), upholding the previously known mechanism of POU5F1 switching partner TFs and binding motifs with different syntax. ( A ) POU5F1 partners with SOX2 to bind its canonical motif. In contrast, POU5F1 associates with SOX17 to bind a compressed motif. ( B ) The co-binding patterns discovered in SOX2 and SOX17 datasets correspond to the expected canonical and compressed motifs. One co-binding pattern was discovered downstream of the anchor with either zero or one nucleotide between them. ( C ) The co-binding motif found in the SOX2 datasets was associated with POU5F1 using motif similarity and PPI data (significant motif similarity P -value < 0.05 and PPI combined score > 500). ( D ) Visual representation of the similarity between the discovered co-binding motif and motifs recognized by POU5F1 in JASPAR and CIS-BP.
In this proof-of-concept example, we knew a priori that POU5F1 was the binding partner of SOX2 and SOX17. Nevertheless, COBIND discovered de novo the co-binding motif. In other settings, one would not know the TF binding to the co-binding motif discovered. To address this challenge, we combined motif similarity to already known motifs from JASPAR and CIS-BP with protein-protein interaction (PPI) data from the STRING database to infer the TFs potentially binding the motifs revealed by COBIND (Materials and methods). This strategy confirmed that the framework inferred POU5F1 as the binding partner of SOX2 and SOX17 (Figure 3C , D ; Additional file 1: Supplementary Figure S31 ).
COBIND discovers known and novel co-binding patterns in a large-scale analysis across several species
We ran COBIND on 5699 UniBind TFBS datasets associated with 401 unique TFs from seven species (Materials and methods; Additional file 1: Supplementary Table S2 ). Beyond predicting co-binding patterns with COBIND, we inferred the TFs likely binding to the discovered patterns following the strategy described above (also see Materials and Methods). Altogether, COBIND revealed 591 co-binding patterns for 224 unique TFs (22 in A. thaliana datasets, 7 in C. elegans , 3 in D. rerio , 22 in D. melanogaster , 309 in H. sapiens , 217 in M. musculus and 11 in R. norvegicus ). For 78% of the reported co-binding motifs (462 out of 591), we found motif similarity and PPI data to support the inferred pair of co-binding TFs (Figure 4 ). Specifically, the data supported all co-binding patterns for D. rerio and C. elegans , more than half of the patterns for R. norvegicus , H. sapiens and M. musculus , 18.2% for A. thaliana and 31.8% for D. melanogaster . We provide all predictions for the community to explore through a dedicated website at https://cbgr.bitbucket.io/COBIND_results_page/ .

Overview of COBIND predicted co-binding patterns. The bar plot provides the number of co-binding patterns (y-axis) discovered by COBIND with (dark pink) or without (light pink) support from motif similarity and PPI data. The bars contain the corresponding numbers and percentages. Each bar summarizes the results for a species (x-axis).
Next, we evaluated the frequency at which a TF A is predicted as a partner of TF B and, conversely, TF B as a partner of TF A . We analyzed co-binding patterns predicted in a specific cell type for each TF A , provided data was available for a predicted co-binding TF B in the same cell type (noting that multiple TFs can recognize the same co-binding pattern). When examining datasets linked to TF B , COBIND successfully predicted back TF A in 77% of the co-binding patterns from human datasets and 84% from mouse datasets.
Furthermore, the analysis of motif similarity between COBIND’s predictions revealed that 67% (143 out of 214) of the TFs shared a co-binding motif with another member of the same TF structural family (Materials and Methods), which confirmed the conservation of co-binding patterns within families of TFs across species. It is noteworthy that anchor regions bound by TFs from the same structural family tend to overlap (Mann–Whitney U test P -value < 0.05) with those bound by different TF families, even though they typically show low Jaccard index values (Additional file 1: Supplementary Figure S32 ). This observation was consistent across different species, covering all regions with and without predicted co-binding patterns (Additional file 1: Supplementary Figure S32 ). However, no statistically significant differences were noted (Mann–Whitney U test P -value > 0.05) when comparing regions with predicted co-binding patterns to those without, irrespective of whether the same or different TF family members bind them.
As a case example, COBIND predicted two co-binding patterns around TEAD1 TFBSs. Specifically, COBIND revealed the same motif with different spacings from the provided anchor TFBSs (2nt upstream or 3nt downstream of TEAD1 TFBSs; Additional file 1: Supplementary Figure S33A ). We found that the two co-binding patterns occurred in ∼6% (3-bp downstream pattern) and ∼7% (2-bp upstream pattern) of the input sequences from mouse nerve tumor cells and in ∼3% (2-bp upstream and 3-bp downstream patterns each) of the sequences from human pancreas and astrocytoma cells. Our approach inferred that TEAD2 and TEAD4 cooperate with TEAD1 (Additional file 1: Supplementary Figure S33B, C ). Previous studies reported that TEAD TFs bind either an isolated M-CAT element or direct DNA repeats with spacing ranging from 0 to 6 bp ( 55–57 ). These studies further support the co-binding patterns predicted by COBIND. The predictions around TEAD1 TFBSs provide an example of COBIND’s capacity to predict co-binding events supported by motif similarity and PPI data.
To exemplify predictions of new co-binding TFs, COBIND revealed a co-binding motif located 4 bp upstream of HY5 anchor TFBSs in A. thaliana (Additional file 1: Supplementary Figure S34A ). The analyses of TFBS datasets associated with the TFs ABF1, ABF3, ABF4, GBF2 and GBF3 displayed the same co-binding pattern. We found that the canonical motifs recognized by human NF-Y TFs (NF-YC and NF-YA) were similar to the co-binding motif discovered by COBIND (Additional file 1: Supplementary Figure S34B, C ). However, no PPI data currently support the physical interactions between the anchor TFs and NF-Y orthologs in plants. Nevertheless, the bZIP67 TF, which belongs to the same basic leucine zipper (bZIP) family as HY5, ABF1, ABF3, ABF4, GBF2 and GBF3, is known to form a transcriptional complex together with NF-YC2 and bind ER stress response elements (ERSEs) to regulate omega-3 fatty acid content in A. thaliana ( 58 ). In agreement with this observation, we noted that the anchor motif combined with the co-binding motif discovered by COBIND forms the motif associated with ERSEs ( 59 ). Furthermore, HY5 competes with bZIP28, another member of the bZIP family, to bind ERSEs on promoters of unfolded protein response genes, and bZIP28 interacts with NF-Y protein complexes ( 59 , 60 ). Consistent with this knowledge, we found that most of the genomic regions predicted by COBIND to harbor the co-binding events in the HY5 dataset were in promoter regions (Additional file 1: Supplementary Figure S34D ). Altogether, these multiple lines of evidence strongly support the co-binding patterns predicted by COBIND between bZIP and NF-Y TFs in A. thaliana .
COBIND discovers the extended motif bound by CTCF
We investigated another example where COBIND predicted a co-binding pattern unsupported by motif similarity and PPI data. Specifically, COBIND identified a co-binding motif in the proximity of CTCF TFBSs for several datasets (Figure 5A ). We observed two spacing configurations between the anchor CTCF TFBSs and the predicted co-binding events. Across the datasets, COBIND predicted from 2.6 to 5% of the sequences to harbor the co-binding patterns. All datasets from human and zebrafish and 25% of the mouse datasets (95 out of 382), which cover a large spectrum of cell types (Additional file 1: Supplementary Table S4 ), contained the identified co-binding patterns (Additional file 1: Supplementary Figure S35 ). Previous studies have reported the motif predicted by COBIND as an extension of the canonical CTCF motif ( 61 , 62 ). These studies revealed that the eighth zinc finger of CTCF acts as a linker instead of a clamp to allow zinc fingers 9–11 to bind the extended motif (Figure 5B ), affecting the binding efficiency, residence time, and binding off-rate of CTCF ( 61 , 62 ). Consequently, COBIND did not predict co-binding between distinct TFs, but CTCF seldomly bound an extended motif through different combinations of zinc finger contacts with the DNA.

CTCF binds a conserved extended motif. ( A ) COBIND predicted two binding configurations through an extended motif for CTCF in human and mouse datasets. The extended binding pattern was discovered upstream of the anchor with either thirteen or fourteen nucleotides between them. ( B ) The two binding configurations derive from contacts between the zinc fingers (ZF) 9–11 and the DNA at the upstream motif. ( C ) Comparison between vertebrate evolutionary conservation of H. sapiens genomic regions harbouring the CTCF canonical motif only (left) and regions with the extended motif revealed higher conservation of the extended motif. The purple lines provide the mean conservation score across the regions considered. The grey lines provide the mean conservation score across the same number of random genomic regions in the human genome. ** indicates a Wilcoxon test P -value < 0.001 at the anchor (red) and motif sites (green).
Since evolutionary conservation is a hallmark of functional importance ( 47 ), we assessed the functional relevance of the genomic regions harboring the discovered binding patterns by examining their evolutionary conservation across vertebrates (Materials and Methods). We observed that the anchor and extended motif sites (Figure 5C , right, red and green segments, respectively) were more conserved in the regions predicted to harbor the extended motif than in the other regions (Wilcoxon test P -value < 0.001 for both segments; Figure 5C , right versus left). The increased evolutionary conservation was consistent across all spacing configurations observed in human and mouse. These results exemplify how COBIND can predict relevant DNA patterns that do not correspond to the co-binding of two proteins but to binding variants for the same TF, a particular case of the zinc finger families.
Genomic regions harboring a co-binding pattern are evolutionarily more conserved than regions without co-binding
We assessed the functional relevance of all the co-binding patterns discovered by COBIND across species by analyzing their evolutionary conservation (Materials and methods). Specifically, we compared the evolutionary conservation of the anchor and co-binding positions (Figure 5C , red and green, respectively) in genomic regions harboring a co-binding pattern, named co-bound regions for simplicity, to those without a predicted co-binding pattern. We first describe the analyses of the case studies presented above as examples. We found that the co-bound regions associated with SOX2::POU5F1 and TEAD1::TEAD in the human and mouse genomes exhibited a significantly increased evolutionary conservation compared to those without predicted co-binding (Wilcoxon test P -value < 0.001; Figure 6A ; Additional file 1: Supplementary Figure S36A ). Notably, we observed increased conservation at the co-binding and the anchor motifs. When considering the co-binding pattern associated with HY5 in A. thaliana , we did not observe a significant difference in conservation between the regions with and without the co-binding pattern. Nevertheless, both the anchor and the co-binding motifs exhibited peaks of evolutionary conservation, confirming the likely functional relevance of the predicted co-binding pattern across the flowering plant kingdom (Additional file 1: Supplementary Figure S37A ).

Analysis of the evolutionary conservation of co-binding patterns discovered by COBIND in the human genome. ( A ) Comparison between vertebrate evolutionary conservation of genomic regions where COBIND predicted a co-binding pattern (right) or not (left). The purple lines provide the mean conservation score across the regions considered. The grey lines provide the mean conservation score across the same number of random genomic regions in the human genome. ** indicates a Wilcoxon test P -value < 0.001. The anchor SOX2 anchor motif is underlined in red, and the co-binding motif is underlined in green. ( B ) We compared the evolutionary conservation scores at the anchors' motif locations in genomic regions harboring a co-binding pattern or not. The left panel represents the histogram of the proportion of datasets with the anchor sites harboring higher evolutionary conservation in co-bound regions than in other regions. The right panel represents the corresponding histogram when comparing genomic locations of the co-binding motif. Dark pink represents observed values, and light pink the expected values computed from random assignment of genomic regions predicted as co-bound or not.
Across all datasets and species, we systematically compared the evolutionary conservation at locations of the anchor and predicted co-binding sites discovered by COBIND in co-bound regions versus regions without co-binding sites. Altogether, we observed across species that 23% (for A. thaliana ), 55% (for D. melanogaster ), 60% (for C. elegans ), 82% (for H. Sapiens ), and 83% (for M. musculus ) of the datasets with predicted co-binding patterns exhibited a co-binding motif with significantly more conservation in co-bound regions than in the other regions (Figure 6B ; Additional file 1: Supplementary Figure S36B - S38 ). Furthermore, the associated anchor sites were more conserved in the co-bound regions for 31% ( A. thaliana ), 55% (for D. melanogaster ), 60% (for C. elegans ), 64% (for M. musculus ), and 67% ( H. sapiens ) of the datasets (Figure 6B ; Additional file 1: Supplementary Figure S36B , Supplementary Figure S37B , Supplementary Figure S38 ). Finally, we observed for 64% of the human datasets that the locations of both the anchor and a co-binding motif were more conserved in the co-bound regions than in regions without co-binding sites (61% for M. musculus , 60% for C. elegans , 45% for D. melanogaster , and 23% for A. thaliana ). We compared with random expectation to further support the higher conservation of the anchor and co-binding sites in co-bound regions than similar locations in regions without a predicted co-binding pattern. Specifically, we randomly assigned, for each dataset, each genomic region to harbor or not a co-binding pattern (keeping the same number of regions in each category as the number predicted by COBIND). Figure 6B and Additional file 1: Supplementary Figures S36B and S37B confirm that more datasets exhibit increased conservation in co-bound regions than expected by chance. The consistent increased evolutionary conservation of the co-binding patterns supports the functional importance of COBIND’s predictions across species. Furthermore, our results suggest that the underlying genomic regions harboring a fixed binding motif syntax are evolutionarily important.
DNase I hypersensitive footprints support the predicted co-binding patterns
We further assessed the co-binding patterns discovered by COBIND by analyzing orthogonal experimental data probing chromatin openness. The DNase-seq assay captures open chromatin regions by revealing DNase I hypersensitive sites (DHS) ( 63 ). Importantly, TFs interacting with the DNA in open chromatin regions protect their TFBSs from the DNase cleavage, which leaves a footprint on the corresponding TFBSs ( 50 , 64 ). We retrieved 66 DHS footprint datasets from 53 human cell types ( 50 ) that matched some of the UniBind TFBS datasets used in this study. This data allowed us to investigate DHS footprints at the discovered co-binding motifs for 70 co-binding patterns associated with 44 TF binding profiles as anchors. For each co-binding pattern, we compared the depth of the DHS footprint at the locations of the co-binding motifs between the co-bound regions and the other regions (Materials and methods).
As we ran COBIND on regions surrounding TFBSs predicted as high-quality direct TF–DNA interactions with ChIP-seq and computational evidence from UniBind, we expected DHS footprints at the anchor TFBSs. Indeed, we observed footprints of TF-DNA interactions; for instance, the DHS footprints observed for the CTCF datasets (Figure 7A ). Notably, the DHS footprints at the CTCF anchor locations were significantly deeper in regions predicted with the extended motif than in the other regions (Figure 7A , red segments). Furthermore, the analyses exhibited deep DHS footprints at the location of the co-binding motifs (Figure 7A , green segment on the right compared to the equivalent locations on the left panel). Overall, we found significantly deeper DHS footprints at the locations of a co-binding motif in the predicted co-bound regions than in the other regions for 85% of the co-binding pattern - DHS dataset pairs. In comparison, 26% was expected by chance when randomly assigned the genomic regions as co-bound or not (Figure 7B , right panel). The anchor TFBSs at co-bound regions exhibited deeper DHS footprints than the TFBSs in other regions for 78% of the pairs, while 21% were expected by chance (Figure 7B , left panel). When considering deeper DHS footprints at both the anchor and a co-binding pattern sites, it was observed for 73% of the pairs, while it was never observed with the random assignments (Figure 7B ). As CTCF datasets represented 43% of the total datasets analyzed here, we performed the same analysis, excluding the CTCF datasets. We found that 53% (25% expected by chance) of the co-binding pattern-DHS dataset pairs showed significant DHS footprints at the anchor TFBSs, and 68% (23% expected by chance) exhibited significantly deeper DHS footprints at the co-binding motif locations in co-bound regions than in the other regions. Altogether, 45% of the pairs exhibited deeper DHS footprints at both the co-bound and anchor TFBS locations. Overall, the DHS footprint analyses supported the co-occupancy of the anchor and the co-binding motifs at the co-bound regions.

DHS footprinting analyses at anchor and co-binding locations. ( A ) The plots represent the average DHS score in regions surrounding CTCF TFBSs in neuronal stem cells (purple lines) or at random regions (grey lines). The left plot provides DHS scores at genomic regions where COBIND did not predict the CTCF extended motif; the right plot provides the DHS scores at genomic regions harbouring the CTCF extended motif. ** indicates a Wilcoxon test P -value < 0.001. The anchor CTCF motif is underlined in red and the upstream portion of the extended motif is in green. ( B ) We compared DHS scores at the anchor motif locations in genomic regions harbouring a co-binding pattern or not. The left panel represents the histogram of the datasets where the DHS footprint was deeper in genomic regions harbouring a co-binding pattern. The right panel represents the corresponding histogram when comparing genomic locations of the co-binding motif locations. Dark pink represents observed values, and light pink the expectation computed from randomly assigning regions as containing or not a co-binding pattern.
Single-molecule footprints support co-occupancy at single-molecule resolution for some TFs
The DHS footprinting analysis described above relied on bulk DNase-seq data. Consequently, the results provided average estimations of the co-occupancy of TFs across cells. We aimed to investigate the co-occupancy of TFs at the co-binding patterns predicted by COBIND at single-molecule resolution. To this end, we considered single-molecule footprinting (SMF) data, which probed the co-occupancy of TFs and nucleosomes at single-molecule resolution for accessible genomic regions in mouse embryonic stem cells (mESC) ( 51 ). The SMF data overlapped 1694 regions with predicted co-binding patterns (483 453 reads) and 54 727 regions (14 457 475 reads) not predicted to contain any co-binding pattern for 17 TFs in mESC (Materials and Methods).
For each genomic region predicted by COBIND to contain a co-binding pattern or not, we determined the fractions of molecules in each of the following five states: (i) accessible, (ii) occupied by nucleosomes, (iii) only occupied at the anchor motif, (iv) only occupied at the co-binding motif or (v) co-occupied at both the anchor and the co-binding motifs. This is done by evaluating the binary methylation status of each molecule for each cytosine in and around the anchor and co-binding pattern sites (Figure 8A ). Figure 8B and C illustrates the SMF data analysis at two genomic regions predicted to harbour co-binding events for SOX2 and POU5F1 (Figure 8B ) and the extended CTCF motif (Figure 8C ). We observed footprints of co-occupancy at both the anchor motif and the co-binding motif locations. Specifically, the analysis of 273 molecules across five replicates revealed co-occupancy for 27% of the molecules when considering the SOX2-associated region (Figure 8B ). Five hundred ninety-two molecules across six replicates revealed co-occupancy of the extended CTCF motif for 59% of the molecules (Figure 8C ). In contrast, the example regions where COBIND did not predict a co-binding pattern for SOX2 or CTCF, we only observed occupancy for the SOX2 anchor (17% of 89 molecules) or the canonical CTCF sites (46% of 297 molecules) while co-binding sites were unoccupied (Additional file 1: Supplementary Figure S39A-B ). When considering all the regions with SMF data, we found a larger fraction of co-occupied anchor and co-binding motif locations on the same molecule in COBIND-predicted co-bound regions than in other regions ( P -value < 0.001; Additional file 1: Supplementary Figure S39C ). Significant differences existed in all state abundances, except for the specific occupancy of the co-binding sites. This agrees with these sites not being occupied when the anchor sites are not either. However, not all anchor TF binding profiles satisfied this observation (Figure 8D ). Moreover, we found that genomic regions predicted by COBIND to harbour a co-binding pattern were more occupied (for ‘Anchor + Co-binding’ occupancy state independently) than the other genomic regions for 13 out of 17 anchor TF; statistical significance was observed for 8 TF ( P -value < 0.05; Figure 8D ). Altogether, the results confirmed that SMF data supported the COBIND co-occupancy predictions for some TFs, with the co-bound regions more accessible or co-occupied at single-molecule resolution.

Single-molecule footprinting analysis. ( A ) For each region, single molecules are grouped and assigned to one of the five states (‘Co-binding + Anchor’, ‘Co-binding’, ‘Anchor’, ‘Accessible’, and ‘Nucleosome’) based on the binary cytosine methylation status in and around anchor and co-binding patterns (for two regions of interest, four bins are analysed). (B, C) Single-locus examples of predicted co-bound regions where the anchor TFBSs associate with SOX2 ( B ) and CTCF ( C ). Line plots in the top panels represent the average methylation levels. We provide logos and locations of the anchor motifs (red) and the predicted co-binding motifs (green). Stacks of sorted single molecules are visualized in the lower panels (light grey indicates methylated Cs, accessible positions; black - unmethylated Cs, protected). The y-axis corresponds to the total number of molecules. Coloured bars and percentages show the proportion of molecules in each state (colours corresponding to A). ( D ) We provide boxplots and dotplots showing the distribution of ‘Anchor + Co-binding’ state frequencies of molecules that overlap the regions in two groups: one with predicted co-binding (shown in dark pink) and one with only an anchor TFBS (shown in light pink). The regions with co-binding events show a significantly higher frequency of molecules as co-occupied (‘Anchor + Co-binding’). The differences between groups persist for molecules in other states. ** represents a Wilcoxon test P -value < 0.05.
We introduced COBIND, a computational framework for discovering de novo space-fixed DNA motif patterns in the vicinity of predetermined genomic regions. We applied COBIND to sets of TFBSs from UniBind to identify locally enriched DNA motifs that define co-binding patterns of cooperative TFs. The method uncovered both established and new co-binding patterns, with most TFs sharing a co-binding motif with other TFs from the same family. Additionally, we inferred, when possible, the TFs most likely co-binding the discovered patterns. We make the collection of co-binding patterns revealed by COBIND available to the scientific community for each of the 214 TFs and 58 TF families across seven species. The co-binding events captured by COBIND are likely functionally relevant since they exhibit higher evolutionary conservation than isolated TFBSs. Furthermore, chromatin and single-molecule footprinting data support the occurrence of the identified co-binding events on the same DNA molecules.
We used a combination of motif similarity and prior knowledge of PPIs to identify TFs that may cooperatively bind to the patterns discovered by COBIND. Identifying well-known TF co-binding events, such as POU5F1 with SOX2 or SOX17, essential pluripotency regulators ( 15 , 16 , 65 ), confirmed the approach's efficacy. Importantly, we used a multi-species collection of DNA binding motifs to predict new co-binding configurations for pairs of TFs currently unsupported by PPIs. We discovered that NF-Y proteins are potential partners for HY5, ABF1, 3–4 and GBF2-3 TFs. Previous studies identified NF-Y proteins as co-binders for bZIP67 and bZIP28 proteins belonging to the same TF family ( 58–60 ). More specifically, other studies also pointed to physical interactions of either HY5 or ABF1, 3–4 with NF-YC9 proteins ( 66 , 67 ). We identified the NF-Y-bound co-binding motifs using motifs associated with human TFs because DNA binding profiles for NF-Y are limited in A. thaliana . As HY5 competes with bZIP28, we acknowledge that the NF-Y binding pattern observed close to HY5 TFBSs might be occupied only when bZIP28 is binding. Nevertheless, using multi-species libraries generated potential co-binding predictions that will require further validations to decipher the binding cooperation of TFs at the corresponding genomic regions.
We observed that the COBIND predictions associated with CTCF anchor TFBSs exhibited a binding pattern that did not result from the co-binding of two TFs. Previous studies have discussed this binding pattern and have suggested that CTCF co-binds with ZBTB3 in mouse liver and various human cell lines, including embryonic stem cells and K562 ( 68 , 69 ). Based on motif similarity, we inferred that ZBTB3 could also bind the additional motif identified by COBIND. Instead, the co-binding pattern identified by COBIND corresponds to an extended binding motif for CTCF, involving zinc fingers 9–11 ( 61 ). This orthogonal evidence allowed the two variants of extended CTCF motifs to complement the JASPAR database (MA1930.1 and MA1929.1) ( 7 , 70 ). Furthermore, this binding pattern is critical for optimal CTCF protein residence time on DNA ( 61 ). Importantly, we found that the genomic regions harbouring the extended motif were conserved in both mouse and human genomes, supporting the functional relevance of this binding pattern. Studies frequently reported that zinc-finger proteins bind DNA through a subset of their zinc fingers, but it remains unclear whether the other fingers have additional binding preferences ( 71 ). Thus, binding to extended motifs with additional zinc fingers may be a common characteristic of zinc finger proteins that de novo motif prediction tools like COBIND could capture.
The conservation of TFBSs is an essential indicator of their functional relevance in gene regulation ( 47 ). Furthermore, the combinatorial binding of TFs is important for evolutionary stability, and increased co-binding of TFs associates with a higher probability of a regulatory region being conserved ( 72 ). This study analysed the conservation of co-binding patterns across different species. The regions predicted to harbor TFBSs with fixed spacing were more evolutionarily conserved than those with a single TFBS. The increase in conservation at regions where multiple TFs are likely co-binding compared to single events confirms other reports showing that (i) TFBSs of cooperative TFs are more evolutionarily conserved in mammalian embryonic stem cells ( 73 ) and drosophila ( 74 ) and (ii) that TF cooperativity can increase speed of evolution ( 75 ). Interestingly, not only were the predicted co-bound motif locations conserved, but the anchor motif locations also showed an increase in conservation, which agrees with the previous reports. We found most co-binding patterns with significant conservation from the human and mouse datasets, which may reflect the more significant number of datasets analysed for these species. Overall, the increased conservation of both the anchor and co-binding sites was found in more datasets than expected by chance. However, this was not observed for A. thaliana ; we hypothesize that this is due to the limited number of conservation tracks, which was restricted to flowering plants ( 48 ).
Complementing the conservation analysis, similar results were observed in the DHS footprinting data. Predicted co-binding patterns, along with their anchors, exhibited deeper footprints compared to single TFBSs. A lower signal may indicate a higher false positive rate for single binding events. However, it can also reflect single binding events occurring in a smaller subset of cells while cooperative binding would be shared across cells. Future experiments probing TF binding at single-cell resolution would allow for assessing this hypothesis.
COBIND is a tool for the de novo discovery of co-occurring DNA patterns. Our synthetic data assessment found that STREME performed well in retrieving the injected motifs de novo . However, it underperformed with small numbers of sequences. Moreover, STREME, unlike COBIND, is not intended to capture specific cooperative binding events; therefore, capturing the specific spacings and orientations relative to an anchor motif would require specific post-processing steps. Importantly, these tools do not require a priori knowledge of DNA motifs, such as a known DNA motif binding library, to perform the discovery. The requirement of known motif collections as an input is a standard limitation for other tools dealing with TF binding analysis, such as SpaMo or TACO ( 20 , 21 ). This reliance on existing collections of DNA binding motifs can be problematic as many TF binding motifs still need to be discovered or included. The only input required for COBIND is a set of genomic reference regions to ‘anchor’ the analysis and generate regions where it will search for patterns. The de novo discovery can reveal potential new or unannotated DNA pattern motifs, thereby circumventing the constraints of other tools. As such, COBIND offers a powerful alternative to other motif discovery tools that rely on existing collections of DNA binding motifs. Nonetheless, a limitation is that additional analysis will be required to interpret newly discovered motifs.
In this study, we applied COBIND to genomic regions near TFBSs. However, one can use COBIND in different biological contexts. As a test case, we generated anchor regions for analysis by taking two nucleotides at the donor and acceptor sites of intron-exon boundaries in the human genome. Analysis with COBIND recovered known donor and acceptor motifs (Additional file 2: Supplementary Figure S40 ), demonstrating the versatility of the approach for DNA pattern discovery in other types of biological problems.
We utilized the available SMF data to assess the co-occupancy of the anchor and co-binding motif instances on the same DNA molecules ( 52 ). A limitation of the assay is that one can only apply it to organisms and cell lines that can survive without endogenous methylation. We restricted our analyses to mouse ESCs, for which SMF was already available. A recent study assessed chromatin openness using methylation in the GpC context, allowing applications to different cell types with larger genomic coverage ( 76 ). The SMF data we used was produced with a targeted sequencing approach. Consequently, only a selected set of genomic regions was analyzed, and the overlap with regions containing COBIND-predicted co-binding patterns was incomplete, limiting the full understanding of possible co-occupancy on single molecules.
We recognize that COBIND and this study have several limitations. A fundamental limitation of COBIND is the restricted prediction of co-binding motifs with fixed spacing with the anchor motif. Because of the strict spacing requirements and the limitations of the NMF, COBIND restricts the search space to the close proximity of TFBSs. Previous studies have hypothesized the existence of two distinct sets of enhancers with distinct information-processing mechanisms ( 10 ). One proposed mechanism distinguishes between enhancers where TF binding is highly cooperative and coordinated with fixed spacing, referred to as the ‘enhanceosome’ model, and enhancers where TF cooperation is flexible, known as the ‘billboard’ model ( 77 ). Regardless, these mechanisms are compatible with the presence of both fixed and flexible cooperation of TFs at regulatory elements, depending on the type of TF cooperation. Recent studies support both mechanisms, and even though some suggest that TFBS spacing and orientation are not key determinants of transcription regulation, other studies contradict this statement ( 10–14 ). The evolutionary conservation of the co-binding patterns revealed by COBIND further argues for the functional relevance of a strict grammar for a set of TFs and binding regions.
COBIND relies on the NMF algorithm to discover the co-binding patterns, so it requires enough sequences harbouring the fixed pattern. Our simulated data showed that we obtained reliable results when at least 10% of at least 800 sequences (also 5% of 1000 sequences) contained the pattern to be predicted by COBIND. Since we considered the anchor TFBS and the flanks independently in the COBIND processing, discovering overlapping motifs represents a challenge. COBIND could detect such overlaps by revealing a small co-binding motif, but interpreting such results becomes increasingly difficult as the partial motif becomes small. Furthermore, we used anchor TFBSs predicted from TF binding profiles corresponding to the canonical motifs recognized by the corresponding TFs. This approach prohibits the identification of co-binding patterns where the anchor TFs would recognize altered motifs when cooperating with other proteins. Furthermore, repetitive and low-complexity regions are established issues for de novo motif discovery tools ( 78 , 79 ). For instance, A/T-rich patterns with low complexity, such as A/T stretches, were discovered in 0.5% of random genomics regions matching the %GC composition of the real datasets used for the DHS analysis (Additional file 2: Supplementary Figure S41 - Supplementary Figure S42 ). Therefore, we recommend that users interpret such patterns cautiously. Finally, another limitation lies in the computational time necessary for motif clustering when the NMF identifies many possible motifs. Nevertheless, our comparison to other tools revealed that COBIND’s computational time was not prohibitive.
COBIND is implemented in Python, R, and C++. The COBIND source code and documentation are available at https://bitbucket.org/CBGR/cobind_tool/src/main/ . The source code and data to reproduce the results outlined in this report are available at https://bitbucket.org/CBGR/cobind_manuscript/src/master/ and pre-processed data deposited at https://doi.org/10.5281/zenodo.7681482 . All results presented here are available to the community through a webpage at https://cbgr.bitbucket.io/COBIND_results_page/ .
Supplementary Data are available at NAR Online.
As ‘research parasites’ ( 80 ), we thank all the researchers who made their data available. We thank Marcel Schulz for his suggestion on protein-protein interaction data analysis, François Parcy for fruitful discussion on the plant co-binding motifs and his help finding the plant evolutionary conservation data, Judith Zaugg for providing feedback on the project, Roza Berhanu Lemma, Vipin Kumar, Ladislav Hovan and Rafael Riudavets Puig for reading the manuscript and providing valuable feedback, Harold Gutch, Torfinn Nome, and the NCMM IT team for their IT support, Ingrid Kjelsvik for administrative support, and the members of the Kuijjer and Mathelier groups for insightful discussions.
We follow here the Contributor Roles Taxonomy (CRediT) ( 81 ). Ieva Rauluseviciute: methodology, software, validation, investigation, formal analysis, writing - original draft, visualization; Timothée Launay: methodology, investigation, software, writing - review & editing; Guido Barzaghi: resources, writing - review & editing; Sarvesh Nikumbh: writing - review & editing; Boris Lenhard: writing - review & editing, funding acquisition; Arnaud Regis Krebs: writing - review & editing, funding acquisition; Jaime A. Castro-Mondragon: conceptualization, methodology, writing - review & editing; Anthony Mathelier: conceptualization, methodology, writing - review & editing, supervision, project administration, funding acquisition.
Research Council of Norway [187615]; Helse Sør-Øst; University of Oslo through the Centre for Molecular Medicine Norway (NCMM) (to Mathelier group); Norwegian Cancer Society [197884 to Mathelier group]; Research Council of Norway [288404 to Mathelier group]; Nordic EMBL Partnership Hub for Molecular Medicine, NordForsk Grant [96782 to I.R.]; Deutsche Forschungsgemeinschaft [KR 5247/1-1, salary of G.B.]; core funding from the EMBL and the Deutsche Forschungsgemeinschaft [KR 5247/1-2] (to support research in the laboratory of A.R.K.); Wellcome Trust Joint-Investigator award [106955/Z/15/Z to B.L.]; core funding from the MRC LMS (salary for S.N.). Funding for open access charge: Research Council of Norway.
Conflict of interest statement . None declared.
Wingender E. , Schoeps T. , Haubrock M. , Krull M. , Dönitz J. TFClass: expanding the classification of human transcription factors to their mammalian orthologs . Nucleic Acids Res. 2018 ; 46 : D343 – D347 .
Google Scholar
Lambert S.A. , Jolma A. , Campitelli L.F. , Das P.K. , Yin Y. , Albu M. , Chen X. , Taipale J. , Hughes T.R. , Weirauch M.T. The Human Transcription Factors . Cell . 2018 ; 172 : 650 – 665 .
Zeitlinger J. Seven myths of how transcription factors read the cis-regulatory code . Curr. Opin. Syst. Biol. 2020 ; 23 : 22 – 31 .
Suter D.M. Transcription factors and DNA play hide and seek . Trends Cell Biol. 2020 ; 30 : 491 – 500 .
Weirauch M.T. , Yang A. , Albu M. , Cote A.G. , Montenegro-Montero A. , Drewe P. , Najafabadi H.S. , Lambert S.A. , Mann I. , Cook K. et al. . Determination and inference of eukaryotic transcription factor sequence specificity . Cell . 2014 ; 158 : 1431 – 1443 .
Kulakovskiy I.V. , Vorontsov I.E. , Yevshin I.S. , Sharipov R.N. , Fedorova A.D. , Rumynskiy E.I. , Medvedeva Y.A. , Magana-Mora A. , Bajic V.B. , Papatsenko D.A. et al. . HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis . Nucleic Acids Res. 2018 ; 46 : D252 – D259 .
Castro-Mondragon J.A. , Riudavets-Puig R. , Rauluseviciute I. , Berhanu Lemma R. , Turchi L. , Blanc-Mathieu R. , Lucas J. , Boddie P. , Khan A. , Manosalva Pérez N. et al. . JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles . Nucleic Acids Res. 2022 ; 50 : D165 – D173 .
Reiter F. , Wienerroither S. , Stark A. Combinatorial function of transcription factors and cofactors . Curr. Opin. Genet. Dev. 2017 ; 43 : 73 – 81 .
Zhou M. , Li H. , Wang X. , Guan Y. Evidence of widespread, independent sequence signature for transcription factor cobinding . Genome Res. 2020 ; 31 : 265 – 278 .
Arnosti D.N. , Kulkarni M.M. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? . J. Cell. Biochem. 2005 ; 94 : 890 – 898 .
King D.M. , Hong C.K.Y. , Shepherdson J.L. , Granas D.M. , Maricque B.B. , Cohen B.A. Synthetic and genomic regulatory elements reveal aspects of cis-regulatory grammar in mouse embryonic stem cells . eLife . 2020 ; 9 : e41279 .
Avsec Ž. , Weilert M. , Shrikumar A. , Krueger S. , Alexandari A. , Dalal K. , Fropf R. , McAnany C. , Gagneur J. , Kundaje A. et al. . Base-resolution models of transcription-factor binding reveal soft motif syntax . Nat. Genet. 2021 ; 53 : 354 – 366 .
Sahu B. , Hartonen T. , Pihlajamaa P. , Wei B. , Dave K. , Zhu F. , Kaasinen E. , Lidschreiber K. , Lidschreiber M. , Daub C.O. et al. . Sequence determinants of human gene regulatory elements . Nat. Genet. 2022 ; 54 : 283 – 294 .
Georgakopoulos-Soares I. , Deng C. , Agarwal V. , Chan C.S.Y. , Zhao J. , Inoue F. , Ahituv N. Transcription factor binding site orientation and order are major drivers of gene regulatory activity . Nat. Commun. 2023 ; 14 : 2333 .
Li M. , Belmonte J.C.I. Deconstructing the pluripotency gene regulatory network . Nat. Cell Biol. 2018 ; 20 : 382 – 392 .
Aksoy I. , Jauch R. , Chen J. , Dyla M. , Divakar U. , Bogu G.K. , Teo R. , Leng Ng C.K. , Herath W. , Lili S. et al. . Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm . EMBO J. 2013 ; 32 : 938 – 953 .
Nagy G. , Nagy L. Motif grammar: the basis of the language of gene expression . Comput. Struct. Biotechnol. J. 2020 ; 18 : 2026 – 2032 .
Jauch R. , Aksoy I. , Hutchins A.P. , Ng C.K.L. , Tian X.F. , Chen J. , Palasingam P. , Robson P. , Stanton L.W. , Kolatkar P.R. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA . Stem Cells . 2011 ; 29 : 940 – 951 .
Jolma A. , Yin Y. , Nitta K.R. , Dave K. , Popov A. , Taipale M. , Enge M. , Kivioja T. , Morgunova E. , Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity . Nature . 2016 ; 534 : S15 – S16 .
Jankowski A. , Prabhakar S. , Tiuryn J. TACO: a general-purpose tool for predicting cell-type-specific transcription factor dimers . Bmc Genomics . 2014 ; 15 : 208 .
Whitington T. , Frith M.C. , Johnson J. , Bailey T.L. Inferring transcription factor complexes from ChIP-seq data . Nucleic Acids Res. 2011 ; 39 : e98 .
Levitsky V. , Zemlyanskaya E. , Oshchepkov D. , Podkolodnaya O. , Ignatieva E. , Grosse I. , Mironova V. , Merkulova T. A single ChIP-seq dataset is sufficient for comprehensive analysis of motifs co-occurrence with MCOT package . Nucleic Acids Res. 2019 ; 47 : e139 .
Bentsen M. , Heger V. , Schultheis H. , Kuenne C. , Looso M. TF-COMB - Discovering grammar of transcription factor binding sites . Comput. Struct. Biotechnol. J. 2022 ; 20 : 4040 – 4051 .
Park P.J. ChIP–seq: advantages and challenges of a maturing technology . Nat. Rev. Genet. 2009 ; 10 : 669 – 680 .
van Helden J. , Rios A.F. , Collado-Vides J. Discovering regulatory elements in non-coding sequences by analysis of spaced dyads . Nucleic Acids Res. 2000 ; 28 : 1808 – 1818 .
Defrance M. , Janky R.’s. , Sand O. , van Helden J. Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences . Nat. Protoc. 2008 ; 3 : 1589 – 1603 .
Minnoye L. , Taskiran I.I. , Mauduit D. , Fazio M. , Van Aerschot L. , Hulselmans G. , Christiaens V. , Makhzami S. , Seltenhammer M. , Karras P. et al. . Cross-species analysis of enhancer logic using deep learning . Genome Res. 2020 ; 30 : 1815 – 1834 .
Stein-O’Brien G.L. , Arora R. , Culhane A.C. , Favorov A.V. , Garmire L.X. , Greene C.S. , Goff L.A. , Li Y. , Ngom A. , Ochs M.F. et al. . Enter the matrix: factorization uncovers knowledge from omics . Trends Genet. 2018 ; 34 : 790 – 805 .
Lee D.D. , Seung H.S. Learning the parts of objects by non-negative matrix factorization . Nature . 1999 ; 401 : 788 – 791 .
Devarajan K. Nonnegative matrix factorization: an analytical and interpretive tool in computational biology . PLoS Comput. Biol. 2008 ; 4 : e1000029 .
Nikumbh S. , Lenhard B. Identifying promoter sequence architectures via a chunking-based algorithm using non-negative matrix factorisation . PLoS Comput. Biol. 2023 ; 19 : e1011491 .
Mölder F. , Jablonski K.P. , Letcher B. , Hall M.B. , Tomkins-Tinch C.H. , Sochat V. , Forster J. , Lee S. , Twardziok S.O. , Kanitz A. et al. . Sustainable data analysis with Snakemake . F1000Res . 2021 ; 10 : 33 .
Puig R.R. , Boddie P. , Khan A. , Castro-Mondragon J.A. , Mathelier A. UniBind: maps of high-confidence direct TF-DNA interactions across nine species . Bmc Genomics . 2021 ; 22 : 482 .
Quinlan A.R. , Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features . Bioinformatics . 2010 ; 26 : 841 – 842 .
Pedregosa F. Scikit-learn: machine learning in Python . J. Mach. Learn. Res. 2011 ; 12 : 2825 – 2830 .
Kim H. , Park H. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis . Bioinformatics . 2007 ; 23 : 1495 – 1502 .
Gini C. Variabilità e mutabilità: contributo allo studio delle distribuzioni e delle relazioni statistiche . 1912 ; [Fasc. I.] Tipogr. di P. Cuppini .
Castro-Mondragon J.A. , Jaeger S. , Thieffry D. , Thomas-Chollier M. , van Helden J. RSAT matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections . Nucleic Acids Res. 2017 ; 45 : e119 .
Shrikumar A. , Tian K. , Avsec Ž. , Shcherbina A. , Banerjee A. , Sharmin M. , Nair S. , Kundaje A. Technical Note on Transcription Factor Motif Discovery from Importance Scores (TF-MoDISco) version 0.5.6.5 . 2020 ; arXiv doi: 30 April 2020, preprint: not peer reviewed https://arxiv.org/abs/1811.00416 .
Gupta S. , Stamatoyannopoulos J.A. , Bailey T.L. , Noble W.S. Quantifying similarity between motifs . Genome Biol. 2007 ; 8 : R24 .
Khan A. , Riudavets Puig R. , Boddie P. , Mathelier A. BiasAway: command-line and web server to generate nucleotide composition-matched DNA background sequences . Bioinformatics . 2020 ; 37 : 1607 – 1609 .
Bailey T.L. STREME: accurate and versatile sequence motif discovery . Bioinformatics . 2021 ; 37 : 2834 – 2840 .
Thomas-Chollier M. , Darbo E. , Herrmann C. , Defrance M. , Thieffry D. , van Helden J. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs . Nat. Protoc. 2012 ; 7 : 1551 – 1568 .
Thomas-Chollier M. , Herrmann C. , Defrance M. , Sand O. , Thieffry D. , van Helden J. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets . Nucleic Acids Res. 2012 ; 40 : e31 .
Fornes O. , Castro-Mondragon J.A. , Khan A. , van der Lee R. , Zhang X. , Richmond P.A. , Modi B.P. , Correard S. , Gheorghe M. , Baranašić D. et al. . JASPAR 2020: update of the open-access database of transcription factor binding profiles . Nucleic Acids Res. 2020 ; 48 : D87 – D92 .
Szklarczyk D. , Gable A.L. , Lyon D. , Junge A. , Wyder S. , Huerta-Cepas J. , Simonovic M. , Doncheva N.T. , Morris J.H. , Bork P. et al. . STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets . Nucleic Acids Res. 2019 ; 47 : D607 – D613 .
Siepel A. , Bejerano G. , Pedersen J.S. , Hinrichs A.S. , Hou M. , Rosenbloom K. , Clawson H. , Spieth J. , Hillier L.W. , Richards S. et al. . Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes . Genome Res. 2005 ; 15 : 1034 – 1050 .
Tian F. , Yang D.-C. , Meng Y.-Q. , Jin J. , Gao G. PlantRegMap: charting functional regulatory maps in plants . Nucleic Acids Res. 2020 ; 48 : D1104 – D1113 .
Pohl A. , Beato M. bwtool: a tool for bigWig files . Bioinformatics . 2014 ; 30 : 1618 – 1619 .
Vierstra J. , Lazar J. , Sandstrom R. , Halow J. , Lee K. , Bates D. , Diegel M. , Dunn D. , Neri F. , Haugen E. et al. . Global reference mapping of human transcription factor footprints . Nature . 2020 ; 583 : 729 – 736 .
Sönmezer C. , Kleinendorst R. , Imanci D. , Barzaghi G. , Villacorta L. , Schübeler D. , Benes V. , Molina N. , Krebs A.R. Molecular co-occupancy identifies transcription factor binding cooperativity in vivo . Mol. Cell . 2021 ; 81 : 255 – 267 .
Kleinendorst R.W.D. , Barzaghi G. , Smith M.L. , Zaugg J.B. , Krebs A.R. Genome-wide quantification of transcription factor binding at single-DNA-molecule resolution using methyl-transferase footprinting . Nat. Protoc. 2021 ; 16 : 5673 – 5706 .
Barzaghi G. , Krebs A. , Smith M. SingleMoleculeFootprinting: analysis tools for Single Molecule Footprinting (SMF) data Bioconductor version: release (3.15) . 2022 ;
Mistri T.K. , Devasia A.G. , Chu L.T. , Ng W.P. , Halbritter F. , Colby D. , Martynoga B. , Tomlinson S.R. , Chambers I. , Robson P. et al. . Selective influence of Sox2 on POU transcription factor binding in embryonic and neural stem cells . EMBO Rep. 2015 ; 16 : 1177 – 1191 .
Jiang S.W. , Desai D. , Khan S. , Eberhardt N.L. Cooperative binding of TEF-1 to repeated GGAATG-related consensus elements with restricted spatial separation and orientation . DNA Cell Biol. 2000 ; 19 : 507 – 514 .
Anbanandam A. , Albarado D.C. , Nguyen C.T. , Halder G. , Gao X. , Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain . Proc. Natl. Acad. Sci. U.S.A. 2006 ; 103 : 17225 – 17230 .
Lee D.-S. , Vonrhein C. , Albarado D. , Raman C.S. , Veeraraghavan S. A potential structural switch for regulating DNA-binding by TEAD transcription factors . J. Mol. Biol. 2016 ; 428 : 2557 – 2568 .
Mendes A. , Kelly A.A. , van Erp H. , Shaw E. , Powers S.J. , Kurup S. , Eastmond P.J. bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating FATTY ACID DESATURASE3 . Plant Cell . 2013 ; 25 : 3104 – 3116 .
Pastor-Cantizano N. , Ko D.K. , Angelos E. , Pu Y. , Brandizzi F. Functional diversification of ER stress responses in Arabidopsis . Trends Biochem. Sci. 2020 ; 45 : 123 – 136 .
Nawkar G.M. , Kang C.H. , Maibam P. , Park J.H. , Jung Y.J. , Chae H.B. , Chi Y.H. , Jung I.J. , Kim W.Y. , Yun D.-J. et al. . HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis . Proc. Natl. Acad. Sci. U.S.A. 2017 ; 114 : 2084 – 2089 .
Soochit W. , Sleutels F. , Stik G. , Bartkuhn M. , Basu S. , Hernandez S.C. , Merzouk S. , Vidal E. , Boers R. , Boers J. et al. . CTCF chromatin residence time controls three-dimensional genome organization, gene expression and DNA methylation in pluripotent cells . Nat. Cell Biol. 2021 ; 23 : 881 – 893 .
Nakahashi H. , Kieffer Kwon K.-R. , Resch W. , Vian L. , Dose M. , Stavreva D. , Hakim O. , Pruett N. , Nelson S. , Yamane A. et al. . A genome-wide map of CTCF multivalency redefines the CTCF code . Cell Rep. 2013 ; 3 : 1678 – 1689 .
Thurman R.E. , Rynes E. , Humbert R. , Vierstra J. , Maurano M.T. , Haugen E. , Sheffield N.C. , Stergachis A.B. , Wang H. , Vernot B. et al. . The accessible chromatin landscape of the human genome . Nature . 2012 ; 489 : 75 – 82 .
Funk C.C. , Casella A.M. , Jung S. , Richards M.A. , Rodriguez A. , Shannon P. , Donovan-Maiye R. , Heavner B. , Chard K. , Xiao Y. et al. . Atlas of transcription factor binding sites from ENCODE DNase hypersensitivity data across 27 tissue types . Cell Rep. 2020 ; 32 : 108029 .
Pan X. , Cang X. , Dan S. , Li J. , Cheng J. , Kang B. , Duan X. , Shen B. , Wang Y.-J. Site-specific disruption of the Oct4/Sox2 protein interaction reveals coordinated mesendodermal differentiation and the epithelial-mesenchymal transition . J. Biol. Chem. 2016 ; 291 : 18353 – 18369 .
Kumimoto R.W. , Siriwardana C.L. , Gayler K.K. , Risinger J.R. , Siefers N. , Holt B.F. 3rd NUCLEAR FACTOR Y transcription factors have both opposing and additive roles in ABA-mediated seed germination . PLoS One . 2013 ; 8 : e59481 .
Myers Z.A. , Kumimoto R.W. , Siriwardana C.L. , Gayler K.K. , Risinger J.R. , Pezzetta D. , Holt B.F. III NUCLEAR FACTOR Y, subunit C (NF-YC) transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana . PLoS Genet. 2016 ; 12 : e1006333 .
Wang A.W. , Wang Y.J. , Zahm A.M. , Morgan A.R. , Wangensteen K.J. , Kaestner K.H. The dynamic chromatin architecture of the regenerating liver . Cell. Mol. Gastroenterol. Hepatol. 2020 ; 9 : 121 – 143 .
Boyle A.P. , Song L. , Lee B.-K. , London D. , Keefe D. , Birney E. , Iyer V.R. , Crawford G.E. , Furey T.S. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells . Genome Res. 2011 ; 21 : 456 – 464 .
Rauluseviciute I. , Riudavets-Puig R. , Blanc-Mathieu R. , Castro-Mondragon J.A. , Ferenc K. , Kumar V. , Lemma R.B. , Lucas J. , Chèneby J. , Baranasic D. et al. . JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles . Nucleic Acids Res. 2024 ; 52 : D174 – D182 .
Schmitges F.W. , Radovani E. , Najafabadi H.S. , Barazandeh M. , Campitelli L.F. , Yin Y. , Jolma A. , Zhong G. , Guo H. , Kanagalingam T. et al. . Multiparameter functional diversity of human C2H2 zinc finger proteins . Genome Res. 2016 ; 26 : 1742 – 1752 .
Stefflova K. , Thybert D. , Wilson M.D. , Streeter I. , Aleksic J. , Karagianni P. , Brazma A. , Adams D.J. , Talianidis I. , Marioni J.C. et al. . Cooperativity and rapid evolution of cobound transcription factors in closely related mammals . Cell . 2013 ; 154 : 530 – 540 .
Göke J. , Jung M. , Behrens S. , Chavez L. , O’Keeffe S. , Timmermann B. , Lehrach H. , Adjaye J. , Vingron M. Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development . PLoS Comput. Biol. 2011 ; 7 : e1002304 .
Jiang P. , Singh M. CCAT: combinatorial Code Analysis Tool for transcriptional regulation . Nucleic Acids Res. 2014 ; 42 : 2833 – 2847 .
Tuğrul M. , Paixão T. , Barton N.H. , Tkačik G. Dynamics of Transcription Factor Binding Site Evolution . PLoS Genet. 2015 ; 11 : e1005639 .
Kreibich E. , Kleinendorst R. , Barzaghi G. , Kaspar S. , Krebs A.R. Single-molecule footprinting identifies context-dependent regulation of enhancers by DNA methylation . Mol. Cell . 2023 ; 83 : 787 – 802 .
Slattery M. , Zhou T. , Yang L. , Dantas Machado A.C. , Gordân R. , Rohs R. Absence of a simple code: how transcription factors read the genome . Trends Biochem. Sci. 2014 ; 39 : 381 – 399 .
Dozmorov M.G. , Adrianto I. , Giles C.B. , Glass E. , Glenn S.B. , Montgomery C. , Sivils K.L. , Olson L.E. , Iwayama T. , Freeman W.M. et al. . Detrimental effects of duplicate reads and low complexity regions on RNA- and ChIP-seq data . BMC Bioinf. 2015 ; 16 : S10 .
Chung D. , Kuan P.F. , Li B. , Sanalkumar R. , Liang K. , Bresnick E.H. , Dewey C. , Keleş S. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-Seq data . PLoS Comput. Biol. 2011 ; 7 : e1002111 .
Longo D.L. , Drazen J.M. Data sharing . N. Engl. J. Med. 2016 ; 374 : 276 – 277 .
Brand A. , Allen L. , Altman M. , Hlava M. , Scott J. Beyond authorship: attribution, contribution, collaboration, and credit . Learn. Publ. 2015 ; 28 : 151 – 155 .
Email alerts
Citing articles via.
- Editorial Board
Affiliations
- Online ISSN 1362-4962
- Print ISSN 0305-1048
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
- Create an Account
Enter your email address to receive your free PDF download.
Please note that by doing so you agree to be added to our monthly email newsletter distribution list.

Sign in to Cureus
Sign up for cureus, email communication and personal data.
By joining Cureus, you agree to our Privacy Policy and Terms of Use .
Download Cureus Media Kit
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Preoperative cone beam computed topography assessment of maxillary sinus variations in dental implant patients.

1. Introduction
2. materials and methods, 2.1. study design, 2.2. image analysis, 2.3. statistical analysis, 4. discussion, 5. conclusions, author contributions, institutional review board statement, informed consent statement, data availability statement, conflicts of interest.
- Pelinsari Lana, J.; Moura Rodrigues Carneiro, P.; de Carvalho Machado, V.; Alencar, E.; de Souza, P.; Ricardo Manzi, F.; Campolina Rebello Horta, M. Anatomic variations and lesions of the maxillary sinus detected in cone beam computed tomography for dental implants. Clin. Oral Implants Res. 2012 , 23 , 1398–1403. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Pauwels, R.; Jacobs, R.; Singer, S.R.; Mupparapu, M. CBCT-based bone quality assessment: Are Hounsfield units applicable? Dentomaxillofacial Radiol. 2015 , 44 , 20140238. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Watanabe, H.; Honda, E.; Tetsumura, A.; Kurabayashi, T. A comparative study for spatial resolution and subjective image characteristics of a multi-slice CT and a cone-beam CT for dental use. Eur. J. Radiol. 2011 , 77 , 397–402. [ Google Scholar ] [ CrossRef ]
- Terlemez, A.; Tassoker, M.; Kizilcakaya, M.; Gulec, M. Comparison of cone-beam computed tomography and panoramic radiography in the evaluation of maxillary sinus pathology related to maxillary posterior teeth: Do apical lesions increase the risk of maxillary sinus pathology? Imaging Sci. Dent. 2019 , 49 , 115–122. [ Google Scholar ] [ CrossRef ]
- Krmpotić-Nemanić, J.; Draf, W.; Helms, J. Surgical Anatomy of Head and Neck ; Springer: Berlin/Heidelberg, Germany, 1988. [ Google Scholar ] [ CrossRef ]
- Shankar, L.; Evans, K.; Marotta, T.R.; Yu, E.; Hawke, M.; Stammberger, H. An Atlas of Imaging of the Paranasal Sinuses ; CRC Press: London, UK, 2021. [ Google Scholar ] [ CrossRef ]
- Şimşek Kaya, G.; Daltaban, Ö.; Kaya, M.; Kocabalkan, B.; Sindel, A.; Akdağ, M. The potential clinical relevance of anatomical structures and variations of the maxillary sinus for planned sinus floor elevation procedures: A retrospective cone beam computed tomography study. Clin. Implant. Dent. Relat. Res. 2019 , 21 , 114–121. [ Google Scholar ] [ CrossRef ]
- Traxler, H.; Windisch, A.; Geyerhofer, U.; Surd, R.; Solar, P.; Firbas, W. Arterial blood supply of the maxillary sinus. Clin. Anat. 1999 , 12 , 417–421. [ Google Scholar ] [ CrossRef ]
- Velasco-Torres, M.; Padial-Molina, M.; Alarcón, J.A.; Ovalle, F.; Catena, A.; Galindo-Moreno, P. Maxillary sinus dimensions with respect to the posterior superior alveolar artery decrease with tooth loss. Implant. Dent. 2016 , 25 , 464–470. [ Google Scholar ] [ CrossRef ]
- Gładysz, M.T.; Kruczała, Z.; Bąk, F.; Ochwat, K. The Role of the Alveolar Antral Artery in Oral and Maxillofacial Surgery: A Comprehensive Review. Transl. Res. Anat. 2024 , 36 , 100309. [ Google Scholar ] [ CrossRef ]
- Iwanaga, J.; Wilson, C.; Lachkar, S.; Tomaszewski, K.A.; Walocha, J.A.; Tubbs, R.S. Clinical anatomy of the maxillary sinus: Application to sinus floor augmentation. Anat. Cell Biol. 2019 , 52 , 17–24. [ Google Scholar ] [ CrossRef ]
- Rostetter, C.; Hungerbühler, A.; Blumer, M.; Rücker, M.; Wagner, M.; Stadlinger, B.; Lübbers, H.T. Cone beam computed tomography evaluation of the artery in the lateral wall of the maxillary sinus: Retrospective analysis of 602 sinuses. Implant. Dent. 2018 , 27 , 434–438. [ Google Scholar ] [ CrossRef ]
- Mardinger, O.; Abba, M.; Hirshberg, A.; Schwartz-Arad, D. Prevalence, diameter and course of the maxillary intraosseous vascular canal with relation to sinus augmentation procedure: A radiographic study. Int. J. Oral Maxillofac. Surg. 2007 , 36 , 735–738. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Varela-Centelles, P.; Loira-Gago, M.; Seoane-Romero, J.M.; Takkouche, B.; Monteiro, L.; Seoane, J. Detection of the posterior superior alveolar artery in the lateral sinus wall using computed tomography/cone beam computed tomography: A prevalence meta-analysis study and systematic review. Int. J. Oral Maxillofac. Surg. 2015 , 44 , 1405–1410. [ Google Scholar ] [ CrossRef ]
- Landis, R.; Koch, G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977 , 33 , 159–174. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Potter, R. Oral Radiology: Principles and Interpretation by Stuart White and Michael Pharoah. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2005 , 99 , 253. [ Google Scholar ] [ CrossRef ]
- Blake, F.A.S.; Blessmann, M.; Pohlenz, P.; Heiland, M. A new imaging modality for intraoperative evaluation of sinus floor augmentation. Int. J. Oral Maxillofac. Surg. 2008 , 37 , 183–185. [ Google Scholar ] [ CrossRef ]
- Amine, K.; Slaoui, S.; Kanice, F.Z.; Kissa, J. Evaluation of maxillary sinus anatomical variations and lesions: A retrospective analysis using cone beam computed tomography. J. Stomatol. Oral Maxillofac. Surg. 2020 , 121 , 484–489. [ Google Scholar ] [ CrossRef ]
- Danesh-Sani, S.A.; Movahed, A.; ElChaar, E.S.; Chong Chan, K.; Amintavakoli, N. Radiographic Evaluation of Maxillary Sinus Lateral Wall and Posterior Superior Alveolar Artery Anatomy: A Cone-Beam Computed Tomographic Study. Clin. Implant. Dent. Relat. Res. 2017 , 19 , 151–160. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Ilgüy, D.; Ilgüy, M.; Dolekoglu, S.; Fisekcioglu, E. Evaluation of the posterior superior alveolar artery and the maxillary sinus with CBCT. Oral Radiology Braz. Oral Res. 2013 , 27 , 431–437. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Shiki, K.; Tanaka, T.; Kito, S.; Wakasugi-Sato, N.; Matsumoto-Takeda, S.; Oda, M.; Nishimura, S.; Morimoto, Y. The significance of cone beam computed tomography for the visualization of anatomical variations and lesions in the maxillary sinus for patients hoping to have dental implant-supported maxillary restorations in a private dental office in Japan. Head. Face Med. 2014 , 10 , 20. [ Google Scholar ] [ CrossRef ]
- Naitoh, M.; Suenaga, Y.; Kondo, S.; Gotoh, K.; Ariji, E. Assessment of Maxillary Sinus Septa Using Cone-Beam Computed Tomography: Etiological Consideration. Clin. Implant. Dent. Relat. Res. 2009 , 11 , e52–e58. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Van Den Bergh, J.P.A.; Ten Bruggenkate, C.M.; Disch, F.J.M.; Tuinzing, D.B. Anatomical aspects of sinus floor elevations. Clin. Oral Implants Res. 2000 , 11 , 256–265. [ Google Scholar ] [ CrossRef ]
- Aimetti, M.; Romagnoli, R.; Ricci, G.; Massei, G. Maxillary sinus elevation: The effect of macrolacerations and microlacerations of the sinus membrane as determined by endoscopy. Int. J. Periodontics Restor. Dent. 2001 , 21 , 581–589. [ Google Scholar ]
- Güncü, G.N.; Yildirim, Y.D.; Wang, H.L.; Tözüm, T.F. Location of posterior superior alveolar artery and evaluation of maxillary sinus anatomy with computerized tomography: A clinical study. Clin. Oral Implant. Res. 2011 , 22 , 1164–1167. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Ritter, L.; Lutz, J.; Neugebauer, J.; Scheer, M.; Dreiseidler, T.; Zinser, M.J.; Mischkowski, R.A. Prevalence of pathologic findings in the maxillary sinus in cone-beam computerized tomography. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2011 , 111 , 634–640. [ Google Scholar ] [ CrossRef ]
- Bong-Hae, C.; Yun-Hoa, J. Prevalence of incidental paranasal sinus opacification in an adult dental population. Koreean J. Oral Maxilofac. Radiol. 2009 , 39 , 191–194. [ Google Scholar ]
- Whyte, A.; Boeddinghaus, R. The maxillary sinus: Physiology, development and imaging anatomy. Dentomaxillofacial Radiol. 2019 , 48 , 20190205. [ Google Scholar ] [ CrossRef ]
- Ta Velli, L.; Borgonovo, A.E.; Re, D.; Maiorana, C. Sinus presurgical evaluation: A literature review and a new classification proposal. Minerva Stomatologica. Ed. Minerva Medica 2017 , 66 , 115–131. [ Google Scholar ] [ CrossRef ]
- Mardinger, O.; Manor, I.; Mijiritsky, E.; Hirshberg, A. Maxillary sinus augmentation in the presence of antral pseudocyst: A clinical approach. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2007 , 103 , 180–184. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Yaman, H.; Yilmaz, S.; Karali, E.; Guclu, E.; Ozturk, O. Evaluation and Management of Antrochoanal Polyps. Clin. Exp. Otorhinolaryngol. 2010 , 3 , 110. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Basma, H.S.; Abou-Arraj, R.V. Management of a Large Artery During Maxillary Sinus Bone Grafting: A Case Report. Clin. Adv. Periodontics 2021 , 11 , 22–26. [ Google Scholar ] [ CrossRef ]
- Al-Ghurabi, Z.H.; Abdulrazaq, S.S. Vascular Precautions before Sinus Lift Procedure. J. Craniofacial Surg. 2018 , 29 , e116–e118. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Wang, L.; Gun, R.; Youssef, A.; Carrau, R.L.; Prevedello, D.M.; Otto, B.A.; Ditzel, L. Anatomical study of critical features on the posterior wall of the maxillary sinus: Clinical implications. Laryngoscope 2014 , 124 , 2451–2455. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Valente, N.A. Anatomical Considerations on the Alveolar Antral Artery as Related to the Sinus Augmentation Surgical Procedure. Clin. Implant. Dent. Relat. Res. 2016 , 18 , 1042–1050. [ Google Scholar ] [ CrossRef ] [ PubMed ]
Click here to enlarge figure
| Anatomic Variation | Frequency | Gender Distribution | p Value | Age Group Distribution | p Value | |
|---|---|---|---|---|---|---|
| Pneumatization | 163 (81.5%) -alveolar 32.5% -multiple sites 67.5% | M: 97 (59.5%) F: 66 (40.5%) | 3.246 0.53 | A: 55 (33.7%) B: 91 (55.8%) C: 17 (10.4%) | 4.304 0.116 | |
| Antral septa | 97 (48.5%) | M: 52 (53.6%) F: 45 (46.4%) | 5.641 0.255 | A:35 (36.1%) B: 52 (53.6%) C: 10 (10.3%) | 1.462 0.481 | |
| Hypoplasia | 13 (6.5%) | M: 7 (53.8%) F: 6 (46.2%) | 0.141 0.842 | A: 7 (53.8%) B: 5 (38.5%) C: 1 (7.7%) | 2.498 0.287 | |
| Exostosis | 3 (1.5%) | M: 2 (66.6%) F: 1 (33.3%) | 0.128 0.597 | A:1 (33.3%) B:2 (66.6%) C:0 | 0.304 0.859 | |
| Pathological factors | ||||||
| Mucosal thickening | >3 mm 81 (40.5%) | M: 40 (49.4%) F:41 (50.6%) | 2.8061 0.063 | A: 1 B: 71 C: 9 | 65.605 <0.001 | |
| ≤3 mm 119 (59.5%) | M:73 (61.3%) F:46 (38.7%) | A: 67 B: 44 C: 8 | ||||
| Polypoid lesion | 35 (17.5%) | M: 23 (65.7%) F: 12 (34.3%) | 1.466 0.153 | A: 20 (57.1%) B: 15 (42.9%) C: 0 | 11.871 <0.001 | |
| Bone thickening | 7 (3.5%) | M: 1 (14.3%) F: 6 (85.7%) | 5.260 0.022 | A: 2 (28.6%) B: 4 (57.1%) C: 1 14.3%) | 0.349 0.840 | |
| Antrolith | 4 (2%) | M: 3 (75%) F: 1 (25%) | 0.568 0.414 | A: 1 (25% B: 2 (50%) C: 1 (25%) | 1.444 0.486 | |
| Sinusal floor discontinuity | 43 (21.5%) | Bone graft 36 (83.7%) | M: 20 (55.5%) F: 16 (44.4%) | 2.679 0.444 | A: 18 (50%) B: 11 (30.6%) C: 7 (19.4%) | 16.515 0.011 |
| Extraction 5 (11.6%) | M: 3 (60%) F: 2 (40%) | A: 2 (40%) B: 3 (60%) C: 0 | ||||
| Unclear reason 2 (4.7%) | M: 0 F: 2 (100%) | A: 0 B: 2 (100% C: 0 | ||||
| Foreign body | 3 (1.5%) | M: 1 (33.3%) F: 2 (66.6%) | 0.665 0.403 | A: 0 B: 2 (66.6%) C: 1 (33.3%) | 3.290 0.193 | |
| Opacifiation | 16 (8%) | M: 7 (43.8%) F: 9 (56.2%) | 1.150 0.208 | A: 6 (37.5%) B: 9 (56.3%) C: 1 (6.2%) | 0.171 0.918 | |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Misăiloaie, A.; Tărăboanță, I.; Budacu, C.C.; Sava, A. Preoperative Cone Beam Computed Topography Assessment of Maxillary Sinus Variations in Dental Implant Patients. Diagnostics 2024 , 14 , 1929. https://doi.org/10.3390/diagnostics14171929
Misăiloaie A, Tărăboanță I, Budacu CC, Sava A. Preoperative Cone Beam Computed Topography Assessment of Maxillary Sinus Variations in Dental Implant Patients. Diagnostics . 2024; 14(17):1929. https://doi.org/10.3390/diagnostics14171929
Misăiloaie, Alexandru, Ionuț Tărăboanță, Cristian Constantin Budacu, and Anca Sava. 2024. "Preoperative Cone Beam Computed Topography Assessment of Maxillary Sinus Variations in Dental Implant Patients" Diagnostics 14, no. 17: 1929. https://doi.org/10.3390/diagnostics14171929
Article Metrics
Further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals
Recent Advances in Diagnosing and Treating Post-Prostatectomy Urinary Incontinence
- Urologic Oncology
- Published: 31 August 2024
Cite this article

- Yunlong Li MM 1 na1 ,
- YingMing Xiao MM 1 na1 ,
- Zhengang Shen MM 1 ,
- ShengKe Yang MD 1 ,
- Zeng Li MM 1 ,
- Hong Liao MM 1 &
- Shukui Zhou PhD, MD 1
Radical prostatectomy and radiotherapy are common first-line treatments for clinically localized prostate cancer. Despite advances in surgical technology and multidisciplinary management, post-prostatectomy urinary incontinence (PPI) remains a common clinical complication. The incidence and duration of PPI are highly heterogeneous, varying considerably between individuals. Post-prostatectomy urinary incontinence may result from a combination of factors, including patient characteristics, lower urinary tract function, and surgical procedures. Physicians often rely on detailed medical history, physical examinations, voiding diaries, pad tests, and questionnaires-based symptoms to identify critical factors and select appropriate treatment options. Post-prostatectomy urinary incontinence treatment can be divided into conservative treatment and surgical interventions, depending on the severity and type of incontinence. Pelvic floor muscle training and lifestyle interventions are commonly conservative strategies. When conservative treatment fails, surgery is frequently recommended, and the artificial urethral sphincter remains the “gold standard” surgical intervention for PPI. This review focuses on the diagnosis and treatment of PPI, based on the most recent clinical research and recommendations of guidelines, including epidemiology and risk factors, diagnostic methods, and treatment strategies, aimed at presenting a comprehensive overview of the latest advances in this field and assisting doctors in providing personalized treatment options for patients with PPI.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Price excludes VAT (USA) Tax calculation will be finalised during checkout.
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Explore related subjects
- Medical Imaging
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin . 2023;73(1):17–48. https://doi.org/10.3322/caac.21763 .
Article PubMed Google Scholar
Lehto US, Tenhola H, Taari K, Aromaa A. Patients’ perceptions of the negative effects following different prostate cancer treatments and the impact on psychological well-being: a nationwide survey. Br J Cancer . 2017;116(7):864–73. https://doi.org/10.1038/bjc.2017.30 .
Article PubMed PubMed Central Google Scholar
Moses KA, Sprenkle PC, Bahler C, et al. NCCN guidelines® insights: prostate cancer early detection, Version 1. 2023. J Natl Compr Cancer Netw . 2023;21(3):236–46. https://doi.org/10.6004/jnccn.2023.0014 .
Article Google Scholar
Mottet N, van den Bergh RCN, Briers E, et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer-2020 update part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol . 2021;79(2):243–62. https://doi.org/10.1016/j.eururo.2020.09.042 .
Article CAS PubMed Google Scholar
Sekhoacha M, Riet K, Motloung P, Gumenku L, Adegoke A, Mashele S. Prostate cancer review: genetics, diagnosis, treatment options, and alternative approaches. Molecules . 2022. https://doi.org/10.3390/molecules27175730 .
Abramowitz MC, Pollack A. Postprostatectomy radiation therapy for prostate cancer. Semin Radiat Oncol . 2008;18(1):15–22. https://doi.org/10.1016/j.semradonc.2007.09.003 .
Thompson IM Jr, Tangen CM, Paradelo J, et al. Adjuvant radiotherapy for pathologically advanced prostate cancer: a randomized clinical trial. JAMA . 2006;296(19):2329–35. https://doi.org/10.1001/jama.296.19.2329 .
Litwin MS, Melmed GY, Nakazon T. Life after radical prostatectomy: a longitudinal study. J Urol . 2001;166(2):587–92.
Gacci M, Simonato A, Masieri L, et al. Urinary and sexual outcomes in long-term (5+ years) prostate cancer disease free survivors after radical prostatectomy. Health Qual Life Outcomes . 2009;7:94. https://doi.org/10.1186/1477-7525-7-94 .
Gacci M, De Nunzio C, Sakalis V, Rieken M, Cornu JN, Gravas S. Latest Evidence on post-prostatectomy urinary incontinence. J Clin Med . 2023. https://doi.org/10.3390/jcm12031190 .
Kasai T, Banno T, Nakamura K, et al. Duration and influencing factors of postoperative urinary incontinence after robot-assisted radical prostatectomy in a Japanese community hospital: a single-center retrospective cohort study. Int J Environ Res Public Health . 2023. https://doi.org/10.3390/ijerph20054085 .
Lamberg H, Shankar PR, Singh K, et al. Preoperative prostate MRI predictors of urinary continence following radical prostatectomy. Radiology . 2022;303(1):99–109. https://doi.org/10.1148/radiol.210500 .
Krambeck AE, DiMarco DS, Rangel LJ, et al. Radical prostatectomy for prostatic adenocarcinoma: a matched comparison of open retropubic and robot-assisted techniques. BJU Int . 2009;103(4):448–53. https://doi.org/10.1111/j.1464-410X.2008.08012.x .
Singh I, Hemal AK. Robotic-assisted radical prostatectomy in 2010. Expert Rev Anticancer Ther . 2010;10(5):671–82. https://doi.org/10.1586/era.10.35 .
Patel VR, Sivaraman A, Coelho RF, et al. Pentafecta: a new concept for reporting outcomes of robot-assisted laparoscopic radical prostatectomy. Eur Urol . 2011;59(5):702–7. https://doi.org/10.1016/j.eururo.2011.01.032 .
Sacco E, Prayer-Galetti T, Pinto F, et al. Urinary incontinence after radical prostatectomy: incidence by definition, risk factors and temporal trend in a large series with a long-term follow-up. BJU Int . 2006;97(6):1234–41. https://doi.org/10.1111/j.1464-410X.2006.06185.x .
Ficarra V, Novara G, Rosen RC, et al. Systematic review and meta-analysis of studies reporting urinary continence recovery after robot-assisted radical prostatectomy. Eur Urol . 2012;62(3):405–17. https://doi.org/10.1016/j.eururo.2012.05.045 .
Montorsi F, Wilson TG, Rosen RC, et al. Best practices in robot-assisted radical prostatectomy: recommendations of the pasadena consensus Panel. Eur Urol . 2012;62(3):368–81. https://doi.org/10.1016/j.eururo.2012.05.057 .
Holze S, Kuntze AS, Mende M, et al. Assessment of different continence definitions in the context of the randomized multicenter prospective LAP-01 trial-Does the best definition change over time? Eur J Med Res . 2024;29(1):58. https://doi.org/10.1186/s40001-024-01662-5 .
Article CAS PubMed PubMed Central Google Scholar
Moretti TBC, Magna LA, Reis LO. Continence criteria of 193 618 patients after open, laparoscopic, and robot-assisted radical prostatectomy. BJU Int . 2023. https://doi.org/10.1111/bju.16180 .
Holze S, Mende M, Healy KV, et al. Comparison of various continence definitions in a large group of patients undergoing radical prostatectomy: a multicentre, prospective study. BMC Urol . 2019;19(1):70. https://doi.org/10.1186/s12894-019-0500-6 .
Nakane A, Kubota H, Noda Y, et al. Improvement in early urinary continence recovery after robotic-assisted radical prostatectomy based on postoperative pelvic anatomic features: a retrospective review. BMC Urol . 2019;19(1):87. https://doi.org/10.1186/s12894-019-0519-8 .
Hammerer P, Huland H. Urodynamic evaluation of changes in urinary control after radical retropubic prostatectomy. J Urol . 1997;157(1):233–6.
Donnellan SM, Duncan HJ, MacGregor RJ, Russell JM. Prospective assessment of incontinence after radical retropubic prostatectomy: objective and subjective analysis. Urology . 1997;49(2):225–30. https://doi.org/10.1016/s0090-4295(96)00451-7 .
Abuelnaga M, Gendy R, Yao M, et al. Impact of definitions on continence outcomes in a series of 1000 robot-assisted radical prostatectomies, time for an internationally agreed definition? BJU Int . 2024. https://doi.org/10.1111/bju.16327 .
Dubbelman Y, Groen J, Wildhagen M, Rikken B, Bosch R. The recovery of urinary continence after radical retropubic prostatectomy: a randomized trial comparing the effect of physiotherapist-guided pelvic floor muscle exercises with guidance by an instruction folder only. BJU Int . 2010;106(4):515–22. https://doi.org/10.1111/j.1464-410X.2010.09159.x .
Rasmussen A, Mouritsen L, Dalgaard A, Frimodt-Møller C. Twenty-four hour pad weighing test: reproducibility and dependency of activity level and fluid intake. Neurourol Urodyn . 1994;13(3):261–5. https://doi.org/10.1002/1520-6777(1994)13:3%3c261::aid-nau1930130308%3e3.0.co;2-t .
Moore KN, Truong V, Estey E, Voaklander DC. Urinary incontinence after radical prostatectomy: can men at risk be identified preoperatively? J Wound Ostomy Cont Nurs . 2007;34(3):270–9. https://doi.org/10.1097/01.Won.0000270821.91694.56 .
d’Altilia N, Mancini V, Falagario UG, et al. A matched-pair analysis after robotic and retropubic radical prostatectomy: a new definition of continence and the impact of different surgical techniques. Cancers . 2022. https://doi.org/10.3390/cancers14184350 .
Jeong SJ, Yeon JS, Lee JK, et al. Development and validation of nomograms to predict the recovery of urinary continence after radical prostatectomy: comparisons between immediate, early, and late continence. World J Urol . 2014;32(2):437–44. https://doi.org/10.1007/s00345-013-1127-y .
Matsushita K, Kent MT, Vickers AJ, et al. Preoperative predictive model of recovery of urinary continence after radical prostatectomy. BJU Int . 2015;116(4):577–83. https://doi.org/10.1111/bju.13087 .
Stanford JL, Feng Z, Hamilton AS, et al. Urinary and sexual function after radical prostatectomy for clinically localized prostate cancer: the prostate cancer outcomes study. JAMA . 2000;283(3):354–60. https://doi.org/10.1001/jama.283.3.354 .
Lardas M, Grivas N, Debray TPA, et al. Patient- and tumour-related prognostic factors for urinary incontinence after radical prostatectomy for nonmetastatic prostate cancer: a systematic review and meta-analysis. Eur Urol Focus . 2022;8(3):674–89. https://doi.org/10.1016/j.euf.2021.04.020 .
Gondoputro W, Thompson J, Evans M, et al. how does age affect urinary continence following robot-assisted radical prostatectomy? A prospective multi-institutional study using independently collected, validated questionnaires. J Urol . 2022;207(5):1048–56. https://doi.org/10.1097/ju.0000000000002391 .
Strasser H, Tiefenthaler M, Steinlechner M, Bartsch G, Konwalinka G. Urinary incontinence in the elderly and age-dependent apoptosis of rhabdosphincter cells. Lancet . 1999;354(9182):918–9. https://doi.org/10.1016/s0140-6736(99)02588-x .
Ahlering TE, Eichel L, Skarecky D. Evaluation of long-term thermal injury using cautery during nerve sparing robotic prostatectomy. Urology . 2008;72(6):1371–4. https://doi.org/10.1016/j.urology.2007.11.101 .
Kadono Y, Ueno S, Kadomoto S, et al. Use of preoperative factors including urodynamic evaluations and nerve-sparing status for predicting urinary continence recovery after robot-assisted radical prostatectomy: nerve-sparing technique contributes to the reduction of postprostatectomy incontinence. Neurourol Urodyn . 2016;35(8):1034–9. https://doi.org/10.1002/nau.22877 .
Mandel P, Kretschmer A, Chandrasekar T, et al. The effect of BMI on clinicopathologic and functional outcomes after open radical prostatectomy. Urol Oncol . 2014;32(3):297–302. https://doi.org/10.1016/j.urolonc.2013.09.005 .
Teber D, Gözen AS, Cresswell J, Canda AE, Yencilek F, Rassweiler J. Prevention and management of ureteral injuries occurring during laparoscopic radical prostatectomy: the Heilbronn experience and a review of the literature. World J Urol . 2009;27(5):613–8. https://doi.org/10.1007/s00345-009-0428-7 .
Noblett KL, Jensen JK, Ostergard DR. The relationship of body mass index to intra-abdominal pressure as measured by multichannel cystometry. Int Urogynecol J Pelvic Floor Dysfunct . 1997;8(6):323–6. https://doi.org/10.1007/bf02765589 .
Wiltz AL, Shikanov S, Eggener SE, et al. Robotic radical prostatectomy in overweight and obese patients: oncological and validated-functional outcomes. Urology . 2009;73(2):316–22. https://doi.org/10.1016/j.urology.2008.08.493 .
Wolin KY, Luly J, Sutcliffe S, Andriole GL, Kibel AS. Risk of urinary incontinence following prostatectomy: the role of physical activity and obesity. J Urol . 2010;183(2):629–33. https://doi.org/10.1016/j.juro.2009.09.082 .
Corona G, Gacci M, Baldi E, Mancina R, Forti G, Maggi M. Androgen deprivation therapy in prostate cancer: focusing on sexual side effects. J Sexual Med . 2012;9(3):887–902. https://doi.org/10.1111/j.1743-6109.2011.02590.x .
Article CAS Google Scholar
Gacci M, Corona G, Apolone G, et al. Influence of serum testosterone on urinary continence and sexual activity in patients undergoing radical prostatectomy for clinically localized prostate cancer. Prostate Cancer Prostatic Dis . 2010;13(2):168–72. https://doi.org/10.1038/pcan.2010.4 .
Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis . 1987;40(5):373–83. https://doi.org/10.1016/0021-9681(87)90171-8 .
Novara G, Ficarra V, D’Elia C, et al. Evaluating urinary continence and preoperative predictors of urinary continence after robot assisted laparoscopic radical prostatectomy. J Urol . 2010;184(3):1028–33. https://doi.org/10.1016/j.juro.2010.04.069 .
Cakmak S, Canda AE, Ener K, Atmaca AF, Altinova S, Balbay MD. Does type 2 diabetes mellitus have an impact on postoperative early, mid-term and late-term urinary continence after robot-assisted radical prostatectomy? J Endourol . 2019;33(3):201–6. https://doi.org/10.1089/end.2018.0822 .
Teber D, Sofikerim M, Ates M, et al. Is type 2 diabetes mellitus a predictive factor for incontinence after laparoscopic radical prostatectomy? A matched pair and multivariate analysis. J Urol . 2010;183(3):1087–91. https://doi.org/10.1016/j.juro.2009.11.033 .
Dell’Oglio P, Stabile A, Gandaglia G, et al. New surgical approaches for clinically high-risk or metastatic prostate cancer. Expert Rev Anticancer Ther . 2017;17(11):1013–31. https://doi.org/10.1080/14737140.2017.1374858 .
Gu Z, Zheng Z, Zhang W, et al. The development and assessment of a predicting nomogram for the recovery of immediate urinary continence following laparoscopic radical prostatectomy. Front Surg . 2022;9:1071093. https://doi.org/10.3389/fsurg.2022.1071093 .
von Bodman C, Matsushita K, Savage C, et al. Recovery of urinary function after radical prostatectomy: predictors of urinary function on preoperative prostate magnetic resonance imaging. J Urol . 2012;187(3):945–50. https://doi.org/10.1016/j.juro.2011.10.143 .
Hakimi AA, Faleck DM, Agalliu I, Rozenblit AM, Chernyak V, Ghavamian R. Preoperative and intraoperative measurements of urethral length as predictors of continence after robot-assisted radical prostatectomy. J Endourol . 2011;25(6):1025–30. https://doi.org/10.1089/end.2010.0692 .
Mizutani Y, Uehara H, Fujisue Y, et al. Urinary continence following laparoscopic radical prostatectomy: association with postoperative membranous urethral length measured using real-time intraoperative transrectal ultrasonography. Oncol Lett . 2012;3(1):181–4. https://doi.org/10.3892/ol.2011.446 .
Sauer M, Tennstedt P, Berliner C, et al. Predictors of short and long term urinary incontinence after radical prostatectomy in prostate MRI: significance and reliability of standardized measurements. Eur J Radiol . 2019;120:108668. https://doi.org/10.1016/j.ejrad.2019.108668 .
Liu L, Zhou S, Song D, et al. The predictors of short and long term urinary continence recovery after laparoscopic radical prostatectomy: a single cancer center report in China. World J Surg Oncol . 2024;22(1):150. https://doi.org/10.1186/s12957-024-03425-2 .
Valenzi FM, Fuschi A, Al Salhi Y, et al. Is early continence recovery related to the length of spared urethra? A prospective multicenter study comparing preoperative MRI and histologic specimen measurements after robotic radical prostatectomy. Eur J Surg Oncol . 2024;50(6):108319. https://doi.org/10.1016/j.ejso.2024.108319 .
Barakat B, Addali M, Hadaschik B, Rehme C, Hijazi S, Zaqout S. Predictors of early continence recovery following radical prostatectomy, including transperineal ultrasound to evaluate the membranous urethra length (CHECK-MUL study). Diagnostics . 2024. https://doi.org/10.3390/diagnostics14080853 .
Song C, Doo CK, Hong JH, Choo MS, Kim CS, Ahn H. Relationship between the integrity of the pelvic floor muscles and early recovery of continence after radical prostatectomy. J Urol . 2007;178(1):208–11. https://doi.org/10.1016/j.juro.2007.03.044 .
Lee CH, Ha HK. Intravesical prostatic protrusion as a predictor of early urinary continence recovery after laparoscopic radical prostatectomy. Int J Urol . 2014;21(7):653–6. https://doi.org/10.1111/iju.12419 .
Yang B, Zhang F, Xiao C, Lu J, Ma L, Huang Y. Impact of preoperative magnetic resonance imaging anatomic features on urinary continence recovery after laparoscopic radical prostatectomy. Urol Int . 2020;104(3–4):239–46. https://doi.org/10.1159/000506021 .
Tutolo M, Rosiello G, Stabile G, et al. The key role of levator ani thickness for early urinary continence recovery in patients undergoing robot-assisted radical prostatectomy: a multi-institutional study. Neurourol Urodynam . 2022;41(7):1563–72. https://doi.org/10.1002/nau.25001 .
Zhang S, Liang C, Qian J, et al. The impact of three different bladder neck reconstruction techniques on urinary continence after laparoscopic radical prostatectomy. J Endourol . 2020;34(6):663–70. https://doi.org/10.1089/end.2020.0064 .
Jacobsen NE, Moore KN, Estey E, Voaklander D. Open versus laparoscopic radical prostatectomy: a prospective comparison of postoperative urinary incontinence rates. J Urol . 2007;177(2):615–9. https://doi.org/10.1016/j.juro.2006.09.022 .
Martínez-Holguín E, Herranz-Amo F, Lledó-García E, et al. Comparison between laparoscopic and open prostatectomy: postoperative urinary continence analysis. Actas Urol Espanolas . 2020;44(8):535–41. https://doi.org/10.1016/j.acuro.2019.10.002 .
Hu JC, Gu X, Lipsitz SR, et al. Comparative effectiveness of minimally invasive vs open radical prostatectomy. JAMA . 2009;302(14):1557–64. https://doi.org/10.1001/jama.2009.1451 .
Novara G, Ficarra V, Rosen RC, et al. Systematic review and meta-analysis of perioperative outcomes and complications after robot-assisted radical prostatectomy. Eur Urol . 2012;62(3):431–52. https://doi.org/10.1016/j.eururo.2012.05.044 .
Rocco B, Coelho RF, Cozzi G, De Lorenzis E, Patel V. Robot-Assisted Radical Prostatectomy. In: SL Best, SY Nakada, editors. Minimally Invasive Urology: An Essential Clinical Guide to Endourology, Laparoscopy, LESS and Robotics. Springer: New York; 2015. p. 49–77.
Chapter Google Scholar
Galfano A, Ascione A, Grimaldi S, Petralia G, Strada E, Bocciardi AM. A new anatomic approach for robot-assisted laparoscopic prostatectomy: a feasibility study for completely intrafascial surgery. Eur Urol . 2010;58(3):457–61. https://doi.org/10.1016/j.eururo.2010.06.008 .
Liu J, Zhang J, Yang Z, et al. Comparison of Retzius-sparing and conventional robot-assisted laparoscopic radical prostatectomy regarding continence and sexual function: an updated meta-analysis. Prostate Cancer Prostatic Dis . 2022;25(1):47–54. https://doi.org/10.1038/s41391-021-00459-5 .
Ku JY, Ha HK. Learning curve of robot-assisted laparoscopic radical prostatectomy for a single experienced surgeon: comparison with simultaneous laparoscopic radical prostatectomy. World J Men’s Health . 2015;33(1):30–5. https://doi.org/10.5534/wjmh.2015.33.1.30 .
Steineck G, Bjartell A, Hugosson J, et al. Degree of preservation of the neurovascular bundles during radical prostatectomy and urinary continence 1 year after surgery. Eur Urol . 2015;67(3):559–68. https://doi.org/10.1016/j.eururo.2014.10.011 .
Palisaar JR, Roghmann F, Brock M, Löppenberg B, Noldus J, von Bodman C. Predictors of short-term recovery of urinary continence after radical prostatectomy. World J Urol . 2015;33(6):771–9. https://doi.org/10.1007/s00345-014-1340-3 .
Xiang P, Du Z, Guan D, et al. Is there any difference in urinary continence between bilateral and unilateral nerve sparing during radical prostatectomy? A systematic review and meta-analysis. World J Surg Oncol . 2024;22(1):66. https://doi.org/10.1186/s12957-024-03340-6 .
Hollabaugh RS Jr, Dmochowski RR, Kneib TG, Steiner MS. Preservation of putative continence nerves during radical retropubic prostatectomy leads to more rapid return of urinary continence. Urology . 1998;51(6):960–7. https://doi.org/10.1016/s0090-4295(98)00128-9 .
Marshall FF. Risk factors for urinary incontinence after radical prostatectomy. J Urol . 1996;156(5):1724. https://doi.org/10.1016/s0022-5347(01)65491-0 .
Begg CB, Riedel ER, Bach PB, et al. Variations in morbidity after radical prostatectomy. N Engl J Med . 2002;346(15):1138–44. https://doi.org/10.1056/NEJMsa011788 .
Van Kampen M, De Weerdt W, Van Poppel H, et al. Prediction of urinary continence following radical prostatectomy. Urol Int . 1998;60(2):80–4. https://doi.org/10.1159/000030216 .
Creta M, Manfredi C, Arcaniolo D, et al. Functional and oncological outcomes after radical prostatectomy in patients with history of surgery for lower urinary tract symptoms related to benign prostatic enlargement: a systematic review with meta-analysis. Prostate Cancer Prostatic Dis . 2023. https://doi.org/10.1038/s41391-023-00678-y .
Shao IH, Chang YH, Hou CM, Lin ZF, Wu CT. Predictors of short-term and long-term incontinence after robot-assisted radical prostatectomy. J Int Med Res . 2018;46(1):421–9. https://doi.org/10.1177/0300060517715396 .
Liu Y, Qin J, Li KP, et al. Perioperative, functional, and oncologic outcomes in patients undergoing robot-assisted radical prostatectomy previous transurethral resection of prostate: a systematic review and meta-analysis of comparative trials. J Robot Surg . 2023;17(4):1271–85. https://doi.org/10.1007/s11701-023-01555-5 .
Gu L, Li Y, Li X, Liu W. Does previous transurethral resection of the prostate negatively influence subsequent robotic-assisted radical prostatectomy in men diagnosed with prostate cancer? A systematic review and meta-analysis. J Robot Surg . 2023;17(4):1299–307. https://doi.org/10.1007/s11701-023-01588-w .
Leyh-Bannurah SR, Liakos N, Oelke M, et al. Perioperative and postoperative outcomes of robot-assisted radical prostatectomy in prostate cancer patients with prior transurethral subvesical deobstruction: results of a high-volume center. J Urol . 2021;206(2):308–18. https://doi.org/10.1097/ju.0000000000001776 .
Loughlin KR, Prasad MM. Post-prostatectomy urinary incontinence: a confluence of 3 factors. J Urol . 2010;183(3):871–7. https://doi.org/10.1016/j.juro.2009.11.011 .
Eastham JA, Kattan MW, Rogers E, et al. Risk factors for urinary incontinence after radical prostatectomy. J Urol . 1996;156(5):1707–13.
Bianco FJ Jr, Scardino PT, Eastham JA. Radical prostatectomy: long-term cancer control and recovery of sexual and urinary function (“trifecta”). Urology . 2005;66(5 Suppl):83–94. https://doi.org/10.1016/j.urology.2005.06.116 .
Winters JC, Appell RA, Rackley RR. Urodynamic findings in postprostatectomy incontinence. Neurourol Urodynam . 1998;17(5):493–8. https://doi.org/10.1002/(sici)1520-6777(1998)17:5%3c493::aid-nau5%3e3.0.co;2-8 .
Groutz A, Blaivas JG, Chaikin DC, Weiss JP, Verhaaren M. The pathophysiology of post-radical prostatectomy incontinence: a clinical and video urodynamic study. J Urol . 2000;163(6):1767–70.
Ficazzola MA, Nitti VW. The etiology of post-radical prostatectomy incontinence and correlation of symptoms with urodynamic findings. J Urol . 1998;160(4):1317–20.
Kadono Y, Nohara T, Kawaguchi S, et al. Investigating the mechanism underlying urinary continence recovery after radical prostatectomy: effectiveness of a longer urethral stump to prevent urinary incontinence. BJU Int . 2018;122(3):456–62. https://doi.org/10.1111/bju.14181 .
Leach GE, Trockman B, Wong A, Hamilton J, Haab F, Zimmern PE. Post-prostatectomy incontinence: urodynamic findings and treatment outcomes. J Urol . 1996;155(4):1256–9. https://doi.org/10.1016/s0022-5347(01)66235-9 .
Goluboff ET, Chang DT, Olsson CA, Kaplan SA. Urodynamics and the etiology of post-prostatectomy urinary incontinence: the initial Columbia experience. J Urol . 1995;153(3 Pt 2):1034–7.
CAS PubMed Google Scholar
Desautel MG, Kapoor R, Badlani GH. Sphincteric incontinence: the primary cause of post-prostatectomy incontinence in patients with prostate cancer. Neurourol Urodynam . 1997;16(3):153–60. https://doi.org/10.1002/(sici)1520-6777(1997)16:3%3c153::aid-nau4%3e3.0.co;2-d .
Giannantoni A, Mearini E, Zucchi A, et al. Bladder and urethral sphincter function after radical retropubic prostatectomy: a prospective long-term study. Eur Urol . 2008;54(3):657–64. https://doi.org/10.1016/j.eururo.2007.10.054 .
Porena M, Mearini E, Mearini L, Vianello A, Giannantoni A. Voiding dysfunction after radical retropubic prostatectomy: more than external urethral sphincter deficiency. Eur Urol . 2007;52(1):38–45. https://doi.org/10.1016/j.eururo.2007.03.051 .
Trofimenko V, Myers JB, Brant WO. Post-prostatectomy incontinence: how common and bothersome is it really? Sex Med Rev . 2017;5(4):536–43. https://doi.org/10.1016/j.sxmr.2017.05.001 .
Thüroff JW, Abrams P, Andersson KE, et al. EAU guidelines on urinary incontinence. Eur Urol . 2011;59(3):387–400. https://doi.org/10.1016/j.eururo.2010.11.021 .
Sandhu JS, Breyer B, Comiter C, et al. Incontinence after prostate treatment: AUA/SUFU guideline. J Urol . 2019;202(2):369–78. https://doi.org/10.1097/ju.0000000000000314 .
Morey AF, Singla N, Carmel M, et al. Standing cough test for evaluation of post-prostatectomy incontinence: a pilot study. Can J Urol . 2017;24(1):8664–9.
PubMed Google Scholar
O’Sullivan R, Karantanis E, Stevermuer TL, Allen W, Moore KH. Definition of mild, moderate and severe incontinence on the 24-hour pad test. BJOG Int J Obstet Gynaecol . 2004;111(8):859–62. https://doi.org/10.1111/j.1471-0528.2004.00211.x .
Soto González M, Da Cuña Carrera I, Lantarón Caeiro EM, Gutiérrez Nieto M, López García S, Ojea Calvo A. Correlation between the 1-hour and 24-hour pad test in the assessment of male patients with post-prostatectomy urinary incontinence. Prog Urol . 2018;28(11):536–41. https://doi.org/10.1016/j.purol.2018.06.011 .
Sato Y, Tanda H, Nakajima H, et al. Simple and reliable predictor of urinary continence after radical prostatectomy: serial measurement of urine loss ratio after catheter removal. Int J Urol . 2014;21(7):647–51. https://doi.org/10.1111/iju.12400 .
Shy M, Fletcher SG. Objective evaluation of overactive bladder: Which surveys should i use? Curr Bladder Dysfunct Rep . 2013;8(1):45–50. https://doi.org/10.1007/s11884-012-0167-2 .
Machioka K, Kadono Y, Naito R, et al. Evaluating urinary incontinence before and after radical prostatectomy using The International Consultation on Incontinence Questionnaire-Short Form. Neurourol Urodynam . 2019;38(2):726–33. https://doi.org/10.1002/nau.23907 .
Ouanes Y, Hermi A, Chaker K, Bibi M, Mrad Daly K, Nouira Y. Impact of urinary incontinence on the quality of life after open retropubic radical prostatectomy. Cureus . 2022;14(8):e28106. https://doi.org/10.7759/cureus.28106 .
Coyne KS, Margolis MK, Hsieh R, Vats V, Chapple CR. Validation of the urinary sensation scale (USS). Neurourol Urodynam . 2011;30(3):360–5. https://doi.org/10.1002/nau.21005 .
Miller JM, Guo Y, Rodseth SB. Cluster analysis of intake, output, and voiding habits collected from diary data. Nurs Res . 2011;60(2):115–23. https://doi.org/10.1097/NNR.0b013e3182097813 .
van Brummen HJ, Heintz AP, van der Vaart CH. The association between overactive bladder symptoms and objective parameters from bladder diary and filling cystometry. Neurourol Urodynam . 2004;23(1):38–42. https://doi.org/10.1002/nau.10162 .
Homma Y, Kakizaki H, Yamaguchi O, et al. Assessment of overactive bladder symptoms: comparison of 3-day bladder diary and the overactive bladder symptoms score. Urology . 2011;77(1):60–4. https://doi.org/10.1016/j.urology.2010.06.044 .
Hennessey DB, Hoag N, Gani J. Impact of bladder dysfunction in the management of post radical prostatectomy stress urinary incontinence-a review. Transl Androl Urol . 2017;6(Suppl 2):S103–11. https://doi.org/10.21037/tau.2017.04.14 .
Rehder P, Gozzi C. Transobturator sling suspension for male urinary incontinence including post-radical prostatectomy. Eur Urol . 2007;52(3):860–6. https://doi.org/10.1016/j.eururo.2007.01.110 .
Soljanik I, Gozzi C, Becker AJ, Stief CG, Bauer RM. Risk factors of treatment failure after retrourethral transobturator male sling. World J Urol . 2012;30(2):201–6. https://doi.org/10.1007/s00345-011-0671-6 .
Bauer RM, Gozzi C, Roosen A, et al. Impact of the “repositioning test” on postoperative outcome of retroluminar transobturator male sling implantation. Urol Int . 2013;90(3):334–8. https://doi.org/10.1159/000347123 .
Filocamo MT, Li Marzi V, Del Popolo G, et al. Effectiveness of early pelvic floor rehabilitation treatment for post-prostatectomy incontinence. Eur Urol . 2005;48(5):734–8. https://doi.org/10.1016/j.eururo.2005.06.004 .
Centemero A, Rigatti L, Giraudo D, et al. Preoperative pelvic floor muscle exercise for early continence after radical prostatectomy: a randomised controlled study. Eur Urol . 2010;57(6):1039–43. https://doi.org/10.1016/j.eururo.2010.02.028 .
Kannan P, Winser SJ, Fung B, Cheing G. Effectiveness of pelvic floor muscle training alone and in combination with biofeedback, electrical stimulation, or both compared to control for urinary incontinence in men following prostatectomy: systematic review and meta-analysis. Phys Ther . 2018;98(11):932–45. https://doi.org/10.1093/ptj/pzy101 .
Zhou L, Chen Y, Yuan X, Zeng L, Zhu J, Zheng J. Preoperative pelvic floor muscle exercise for continence after radical prostatectomy: a systematic review and meta-analysis. Front Public Health . 2023;11:1186067. https://doi.org/10.3389/fpubh.2023.1186067 .
Goode PS, Burgio KL, Johnson TM 2nd, et al. Behavioral therapy with or without biofeedback and pelvic floor electrical stimulation for persistent postprostatectomy incontinence: a randomized controlled trial. JAMA . 2011;305(2):151–9. https://doi.org/10.1001/jama.2010.1972 .
Wille S, Sobottka A, Heidenreich A, Hofmann R. Pelvic floor exercises, electrical stimulation and biofeedback after radical prostatectomy: results of a prospective randomized trial. J Urol . 2003;170(2 Pt 1):490–3. https://doi.org/10.1097/01.ju.0000076141.33973.75 .
Alan C, Eren AE, Ersay AR, Kocoglu H, Basturk G, Demirci E. Efficacy of duloxetine in the early management of urinary continence after radical prostatectomy. Curr Urol . 2015;8(1):43–8. https://doi.org/10.1159/000365688 .
De Nunzio C, Brucker B, Bschleipfer T, et al. Beyond antimuscarinics: a review of pharmacological and interventional options for overactive bladder management in men. Eur Urol . 2021;79(4):492–504. https://doi.org/10.1016/j.eururo.2020.12.032 .
Nam RK, Herschorn S, Loblaw DA, et al. Population based study of long-term rates of surgery for urinary incontinence after radical prostatectomy for prostate cancer. J Urol . 2012;188(2):502–6. https://doi.org/10.1016/j.juro.2012.04.005 .
Mottet N, Boyer C, Chartier-Kastler E, Ben Naoum K, Richard F, Costa P. Artificial urinary sphincter AMS 800 for urinary incontinence after radical prostatectomy: the French experience. Urol Int . 1998;60(Suppl 2):25–9. https://doi.org/10.1159/000056548 .
Sacomani CAR, Zequi SC, Costa WHD, et al. Long-term results of the implantation of the AMS 800 artificial sphincter for post-prostatectomy incontinence: a single-center experience. Int Braz J Urol . 2018;44(1):114–20. https://doi.org/10.1590/s1677-5538.Ibju.2017.0165 .
Walsh IK, Williams SG, Mahendra V, Nambirajan T, Stone AR. Artificial urinary sphincter implantation in the irradiated patient: safety, efficacy and satisfaction. BJU Int . 2002;89(4):364–8. https://doi.org/10.1046/j.1464-4096.2001.01759.x .
Van der Aa F, Drake MJ, Kasyan GR, Petrolekas A, Cornu JN. The artificial urinary sphincter after a quarter of a century: a critical systematic review of its use in male non-neurogenic incontinence. Eur Urol . 2013;63(4):681–9. https://doi.org/10.1016/j.eururo.2012.11.034 .
Chow PM, Agrawal-Patel S, Wood H, Angermeier K. Prior male sling does not affect outcomes of artificial urinary sphincter. BJU Int . 2024;133(5):564–9. https://doi.org/10.1111/bju.16282 .
Munier P, Nicolas M, Tricard T, Droupy S, Costa P, Saussine C. What if artificial urinary sphincter is not possible? Feasibility and effectiveness of ProACT for patients with persistent stress urinary incontinence after radical prostatectomy treated by sling. Neurourol Urodynam . 2020;39(5):1417–22. https://doi.org/10.1002/nau.24355 .
Beaugerie A, Poinard F, Denormandie A, et al. Comparative urodynamic study in cadaver of urethral pressure profilometry between the UroMems artificial urinary sphincter and the AMS800™. Eur Urolol . 2022;81:876.
Comiter C. Surgery for postprostatectomy incontinence: which procedure for which patient? Nat Rev Urol . 2015;12(2):91–9. https://doi.org/10.1038/nrurol.2014.346 .
Meisterhofer K, Herzog S, Strini KA, Sebastianelli L, Bauer R, Dalpiaz O. Male slings for postprostatectomy incontinence: a systematic review and meta-analysis. Eur Urol Focus . 2020;6(3):575–92. https://doi.org/10.1016/j.euf.2019.01.008 .
Del Giudice F, Huang J, Li S, et al. Contemporary trends in the surgical management of urinary incontinence after radical prostatectomy in the United States. Prostate Cancer Prostatic Dis . 2023;26(2):367–73. https://doi.org/10.1038/s41391-022-00558-x .
Abrams P, Constable LD, Cooper D, et al. Outcomes of a noninferiority randomised controlled trial of surgery for men with urodynamic stress incontinence after prostate surgery (MASTER). Eur Urol . 2021;79(6):812–23. https://doi.org/10.1016/j.eururo.2021.01.024 .
Sacco E, Gandi C, Marino F, et al. Artificial urinary sphincter significantly better than fixed sling for moderate post-prostatectomy stress urinary incontinence: a propensity score-matched study. BJU Int . 2021;127(2):229–37. https://doi.org/10.1111/bju.15197 .
Khouri RK Jr, Ortiz NM, Baumgarten AS, et al. Artificial urinary sphincter outperforms sling for moderate male stress urinary incontinence. Urology . 2020;141:168–72. https://doi.org/10.1016/j.urology.2020.03.028 .
Constable L, Abrams P, Cooper D, et al. Synthetic sling or artificial urinary sphincter for men with urodynamic stress incontinence after prostate surgery: the MASTER non-inferiority RCT. Health Technol Assess . 2022;26(36):1–152. https://doi.org/10.3310/tbfz0277 .
Mühlstädt S, Angulo JC, Mohammed N, Schumann A, Fornara P. Complications of the urinary incontinence system ATOMS: description of risk factors and how to prevent these pitfalls. World J Urol . 2020;38(7):1795–803. https://doi.org/10.1007/s00345-019-02962-w .
Angulo JC, Ruiz S, Lozano M, Arance I, Virseda M, Lora D. Systematic review and meta-analysis comparing Adjustable transobturator male system (ATOMS) and male readjustment mechanical external (REMEEX) system for post-prostatectomy incontinence. World J Urol . 2021;39(4):1083–92. https://doi.org/10.1007/s00345-020-03300-1 .
Tricard T, Song QX, Munier P, et al. Adjustable continence therapy (proACT) for the treatment of male stress urinary incontinence post-prostatectomy: a systematic review and meta-analysis (2023 update). World J Urol . 2023;41(7):1793–802. https://doi.org/10.1007/s00345-023-04452-6 .
Choinière R, Violette PD, Morin M, et al. Evaluation of benefits and harms of surgical treatments for post-radical prostatectomy urinary incontinence: a systematic review and meta-analysis. Eur Urol Focus . 2022;8(4):1042–52. https://doi.org/10.1016/j.euf.2021.09.007 .
Larson T, Jhaveri H, Yeung LL. Adjustable continence therapy (ProACT) for the treatment of male stress urinary incontinence: a systematic review and meta-analysis. Neurourol Urodynam . 2019;38(8):2051–9. https://doi.org/10.1002/nau.24135 .
Download references
Acknowledgments
This research was supported by the Natural Science Foundation of Sichuan Science and Technology Agency (No. 2024NSFSC0699).
Author information
Yunlong Li and YingMing Xiao have contributed equally to this work.
Authors and Affiliations
Department of Urology, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, Affiliated Cancer Hospital of University of Electronic Science and Technology of China, Chengdu, China
Yunlong Li MM, YingMing Xiao MM, Zhengang Shen MM, ShengKe Yang MD, Zeng Li MM, Hong Liao MM & Shukui Zhou PhD, MD
You can also search for this author in PubMed Google Scholar
Contributions
Yunlong Li, Shukui Zhou, and Hong Liao conceptualized the article. Yunlong Li and YingMing Xiao were involved in writing the article. Yunlong Li prepared the figures. Shukui Zhou and Hong Liao critically revised the manuscript and figures. All authors read and approved the final manuscript.
Corresponding authors
Correspondence to Hong Liao MM or Shukui Zhou PhD, MD .
Ethics declarations
All authors declare that they have no conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Li, Y., Xiao, Y., Shen, Z. et al. Recent Advances in Diagnosing and Treating Post-Prostatectomy Urinary Incontinence. Ann Surg Oncol (2024). https://doi.org/10.1245/s10434-024-16110-1
Download citation
Received : 15 July 2024
Accepted : 16 August 2024
Published : 31 August 2024
DOI : https://doi.org/10.1245/s10434-024-16110-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Prostate cancer
- Radical prostatectomy
- Post-prostatectomy urinary incontinence
- Risk factors
- Pelvic floor muscle training
- Artificial urethral sphincter
- Find a journal
- Publish with us
- Track your research

IMAGES
VIDEO
COMMENTS
77F. The Critical Success Factor Method: A review and practical example Vanessa A. Cooper RMIT University [email protected] Abstract Since the CSF method was first proposed by John Rockart in 1979, the method has been adopted for numerous research studies in Information Systems (IS). Like many research
Like many research methods, the CSF method has both its supporters and critics. ... J Manage 17(1):99-120, 1991] and by the Critical Success Factors (CSF) method [Rockart, Harv Bus Rev 57(2):81-93 ...
The study comprised a collection of critical success factors and their subfactors where SEM was used to evaluate and validate each critical success factor, relationship, and its subfactors. After conducting the analysis with 212 questionnaires, five critical success factors and subfactors were identified and validated via SEM and influence the ...
We modified the search term into: (ERP OR Enterprise Resource Planning) AND (Post-implementation) AND (CSF OR Critical success factor) AND (failure OR challenge). 3.1.3. Extraction criteria. ... Essential information including the name of the author, journal publisher, publication year, research methods, and CSFs was recorded in an Excel ...
This paper puts forward a systemic methodology to explore Critical Success Factors (CSFs). The common idea about ranking CSFs is to explore the relationship of 'more important', i.e. directly assessing whether success factor A is more important than factor B. This idea is prominent in the prevailing approaches, e.g. the qualitative inquiry, quantitative analysis, and multi-criteria ...
The Critical Success Factor Method: A review and practical example. Since the CSF method was first proposed by John Rockart in 1979, the method has been adopted for numerous research studies in Information Systems (IS). Like many research methods, the CSF method has both its supporters and critics. Almost thirty years on, this paper provides a ...
1 Critical Success Factor Research. According to a study by Esteves (2004), the critical success factors (CSF) approach has been established and popularized over the last 30 years by a number of researchers, particu-larly Rockart (1979). Today, the approach is increasingly used by consultants and IS depart-ments as a means of support to IS ...
Cooper, Vanessa A., "The Critical Success Factor Method: A review and practical example" (2008). CONF-IRM 2008 Proceedings. 53. Since the CSF method was first proposed by John Rockart in 1979, the method has been adopted for numerous research studies in Information Systems (IS). Like many research methods, the CSF method has both its supporters ...
The concept of CSFs (also known as Key Results Areas or KRAs) was first developed by management consultant D. Ronald Daniel, in his article, "Management Information Crisis." [1] John F. Rockart, of MIT's Sloan School of Management, built on and popularized the concept almost two decades later. He defined CSFs as: "The limited number of areas in ...
of the critical success factor method in the field of information systems planning are pre-sented. With regard to enterprise security management and enterprise resiliency, we discuss ... the basis of our research has been granted by permission of the author. SM Operationally Critical Threat, Asset, and Vulnerability Evaluation is a service mark ...
A critical success factor (CSF) is a specific element or activity that is deemed essential for an organization to achieve its mission or goal. In product management, critical success factors are the key actions a product team takes to deliver successful products that solve user problems. Critical success factors are different from critical ...
Step 2: Conduct Market Analysis - Utilize resources like Scenario Planning Guide to understand market trends and competitor strategies. This analysis helps in setting CSFs that are not only relevant but also competitive. Step 3: Define CSFs - Based on the insights gathered, define clear and measurable CSFs.
A multi-factor econometric model can be created for each stage of BPM adoption, which would reconfirm the critical status of the success factors identified by the authors. Future research could also investigate the relationships and interactions between CSFs, depending on the stage of BPM adoption.
The csf method is recommended to perform the evaluation of the information function in the context of evaluative research. key words and phrases: Critical success factors, mis planning, information sys-tems requirements, information systems, evaluative research. Introduction Methods for eliciting information systems requirements for oper-
Critical Success Factor. Several research conducted over the last few decades has demonstrated the significance of critical success factors. Denial was the first to introduce the concept of critical success criteria in 1961. ... The present study uses cross-sectional and quantitative research methods; thus, different methodologies are being ...
The questionnaire was divided into three parts. In the first part Project Managers were asked to rank the factors on the scale from "-3" to "+3" the influence of a given factor on the project success or failure (Exhibit 1), where "-3" means strong influence on the project failure and "+3" means strong influence on the project ...
One method of determining precisely what information is most needed is the "critical success factors" (CSF) method. Introduced in a Harvard Business Review article entitled "Chief Executives Define Their Own Data Needs" (1), the CSF method is now being utilized in a growing number of organizations.
Critical success factors plotted according to their salience to success. Critical success factors plotted according to their significance to success on a natural logarithmic scale so that factors above and to the right are probably significant (p≤0.05). ... APA handbook of research methods in psychology, vol 2: research designs: quantitative ...
International Journal of Methods in Psychiatric Research, 25(3), 220-231. Crossref. PubMed. Google Scholar. Rosenzweig E. D. (2009). A contingent view of e-collaboration and performance in manufacturing. ... (2013). Getting it done: Critical success factors for project managers in virtual work settings. International Journal of Project ...
Critical success factor (CSF) is a management term for an element s necessary for an organization or project to achieve its mission.To achieve their goals they need to be aware of each key success factor (KSF) and the variations between the keys and the different roles key result area (KRA). [1]Main success keys. A CSF is a critical factor or activity that is required for ensuring the success ...
2.1 Critical Success Factor for Agile Teams. Critical success factor is introduced as an approach which detects names and evaluates an organization's performance [13, 14]. The researchers considered critical success factor as factors that ensure accomplishment, satisfaction and bring motivation for individuals with a healthy competitive work ...
The critical success factor method is a means for identifying these important elements of success. It was originally developed to align information technology planning with the strategic direction of an organization. However, in research and fieldwork undertaken by members of the Survivable Enterprise Management (SEM) team at the Software ...
In taking the lead for qual research here at B2B International HQ, I've compiled a list of the 6 critical factors for delivering a successful qualitative research project. Using the Right Methodology. The crux of any research project is using the correct methodology but within qualitative research it can be even more important.
Transcription factor (TF) binding to DNA is critical to transcription regulation. Although the binding properties of numerous individual TFs are ... We present COBIND, a novel method based on non-negative matrix factorization (NMF) to identify TF co-binding patterns automatically. COBIND applies NMF to one-hot encoded regions flanking known TF ...
Embryo transfer is a pivotal procedure in assisted reproductive technologies (ART). Yet, the success of this process hinges on multiple factors, with endometrial receptivity playing a critical role in determining the likelihood of successful implantation. The endometrial receptivity array (ERA) is an advanced diagnostic tool designed to personalize embryo transfer timing by assessing the ...
Critical thinking as one of the key skills for success in the 21st-century has been considered by many scholars in teacher education. This study tries to examine the interaction of critical thinking disposition with two other key characteristics of successful teachers: cognitive flexibility and self-efficacy. To this end, a sample of pre-service English as a Foreign Language (EFL) teachers was ...
This study aimed to evaluate the pathological factors and anatomical variations in the maxillary sinus in patients undergoing dental implant treatment using cone beam computed tomography (CBCT). CBCT, as a key imaging technique in dentistry, offers high-resolution images to assess bone morphology and quality, crucial for preoperative dental implant planning. Material and methods: The study ...
Radical prostatectomy and radiotherapy are common first-line treatments for clinically localized prostate cancer. Despite advances in surgical technology and multidisciplinary management, post-prostatectomy urinary incontinence (PPI) remains a common clinical complication. The incidence and duration of PPI are highly heterogeneous, varying considerably between individuals. Post-prostatectomy ...